Bacterial genome assembly
Fall 2014 BBB talk
Raj Ayyampalayam
Institute of Bioinformatics
Quantitative Biology Consulting Group
Welcome to BBB
- http://qbcg.uga.edu/bbb-tutorials/
- BBB info and archives
- Need help with running jobs on zcluster?
- Contact us (qbcg@uga.edu).
- GACRC Forums: https://forums.gacrc.uga.edu/
- How to run jobs on zcluster?
- https://wiki.gacrc.uga.edu/wiki/Running_Jobs_on_zcluster
- UGA Galaxy: http://qbcg.uga.edu/galaxy-uga/
- Subscribe to qbcg-announce mailing list
- URL: http://goo.gl/k8WG13 enter your name and e-mail address
Bacterial genomes
- Less complicated than larger genomes
- Has the same issues
- Garbage in garbage out
- Better DNA better sequences
- PCR free library perp
- Multiple DNA extraction (different techniques)
- Size ranges from 2MB to 5MB
Sequencing technologies
- Illumina
- MiSeq
- PE 150 - 4.5 GB to 5 GB
- ~ 20 Genomes in a run
- PE 300 - 13 GB to 15 GB
- ~ 40 - 50 Genomes in a run
- PE 150 - 4.5 GB to 5 GB
- NextSeq
- PE 150
- Med output kit
- 30 GB per run
- 50 - 60 Genomes
- 30 GB per run
- High output kit
- 100 GB per run
- 150 Genomes
- 100 GB per run
- Spike-ins?
- Med output kit
- PE 150
- MiSeq
Sequencing technology cont...
- Long reads
- PacBio
- ~ 300 MB to 500 MB of data per SMRTCell
- Reads of length up 30 KB
- PacBio
- Sequencing strategy
- All Illumina
- Single library
- Multiple library
- Illumina and PacBio
- Single Illumina fragment library (50x)
- PacBio library (20x)
- All Illumina
Assembly programs
- A5 pipeline (http://sourceforge.net/projects/ngopt/)
- Short reads only assembler
- SPAdes (http://bioinf.spbau.ru/spades)
- Can use PacBio sub-reads for scaffolding and repeat resolution
- MaSuRCA (http://www.genome.umd.edu/masurca.html)
- Hybrid assembler
- Can user long reads for assembly
- PBcR (http://wgs-assembler.sourceforge.net/wiki/index.php/PacBioToCA)
- Pacbio specific de novo assembler
- With enough coverage can self correct reads
- Can use Illumina reads to correct PacBio reads and then assemble the corrected reads
-
PBJelly (http://sourceforge.net/p/pb-jelly/wiki/Home/)
-
Scaffolds draft assembly using long reads
-
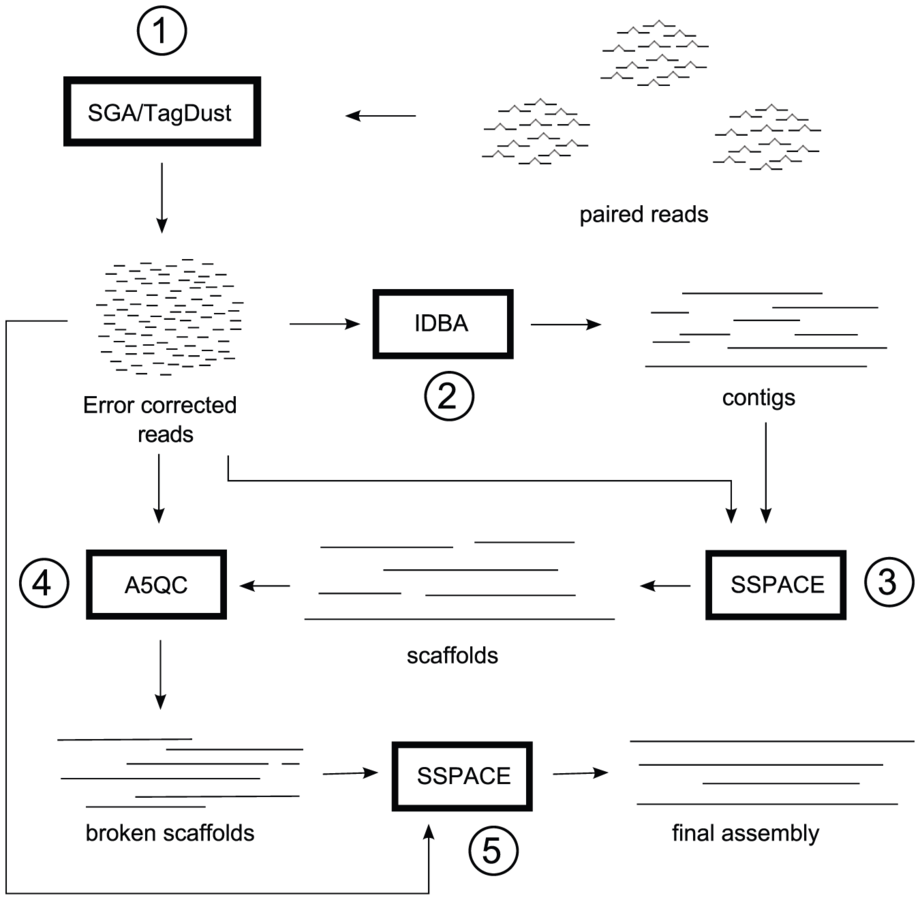
Assembly metrics
- QUAST: Quality Assessment Tool for Genome Assemblies
- http://bioinf.spbau.ru/quast
- Allows for comparison of multiple assemblies at the same time
- Graphical outputs
- Generates “predicted genes” metrics and coordinates
- PHAST: PHAge Seach Tool
- http://phast.wishartlab.com/
- Summarize Assembly
- Part of PBJelly suite
Genome Assemblers Comparision
- GAGE-B http://bioinformatics.oxfordjournals.org/content/29/14/1718.full
- MaSuRCA Best?
- SPAdes analysis using the new version
- http://bioinf.spbau.ru/en/content/spades-30-gage-b-data-sets
- My experience
- Depends on the nature of the genome
- Try different assemblies
- Tune and tweak parameters
- Make your own meta assembly
- http://garm-meta-assem.sourceforge.net/
Questions?Comments
Bacterial Genome Assembly
By Saravanaraj Ayyampalayam
Bacterial Genome Assembly
BBB Presentation - Bacterial genome assembly
- 1,722