Sidney Bell
20 Feb 2019
Center for Viral
Systems Biology
San Diego, CA

Understanding
antigenic evolution
+ resources for
open science

Antigenic variation changes what the virus looks like to your immune system





Measles
Antigenically uniform
Flu
Antigenically
diverse


Lifelong protection
Temporary cross-protection



Influenza & Dengue


Influenza antigenic evolution
Trevor Bedford, Richard Neher & John Huddleston

Population turnover is extremely rapid
Dynamics driven by antigenic drift

Necessitates vaccine updates every ~2 years

Vaccine strain selection by WHO

Which viruses are
antigenically novel?
Measuring antigenic relationships
Measuring antigenic relationships

Titers approximate pairwise
antigenic distance






| 0 | ||
| 0 | 2 | |
| 0 |
1
2



2-fold
change
4-fold
change
4-fold
change
Antigenic Distance
log2(Titer)
Distance
Multidimensional scaling

Antigenic cartography

Smith et al 2004
Source and impact of
antigenic change?
Evolutionary basis of antigenic change
Neher et al, PNAS 2016






| 0 | 1 | 2 |
| 1 | 0 | 2 |
| 2 | 2 | 0 |


Titer
between
&





+ avidity + potency

1.0
0.5
0.5
0.5
Phylogenetic model of
antigenic evolution
Learn these
Minimize
this
Predict titers
Neher et al, PNAS 2016
Phylogenetic model of
antigenic evolution
dTiter on each branch
Virus
avidity
Serum
potency
Predict titers
Neher et al, PNAS 2016
Phylogenetic model of
antigenic evolution
Interpolate across the tree
to estimate antigenic relationships

Neher et al, PNAS 2016
Interpolate across the tree
to estimate antigenic relationships

Neher et al, PNAS 2016
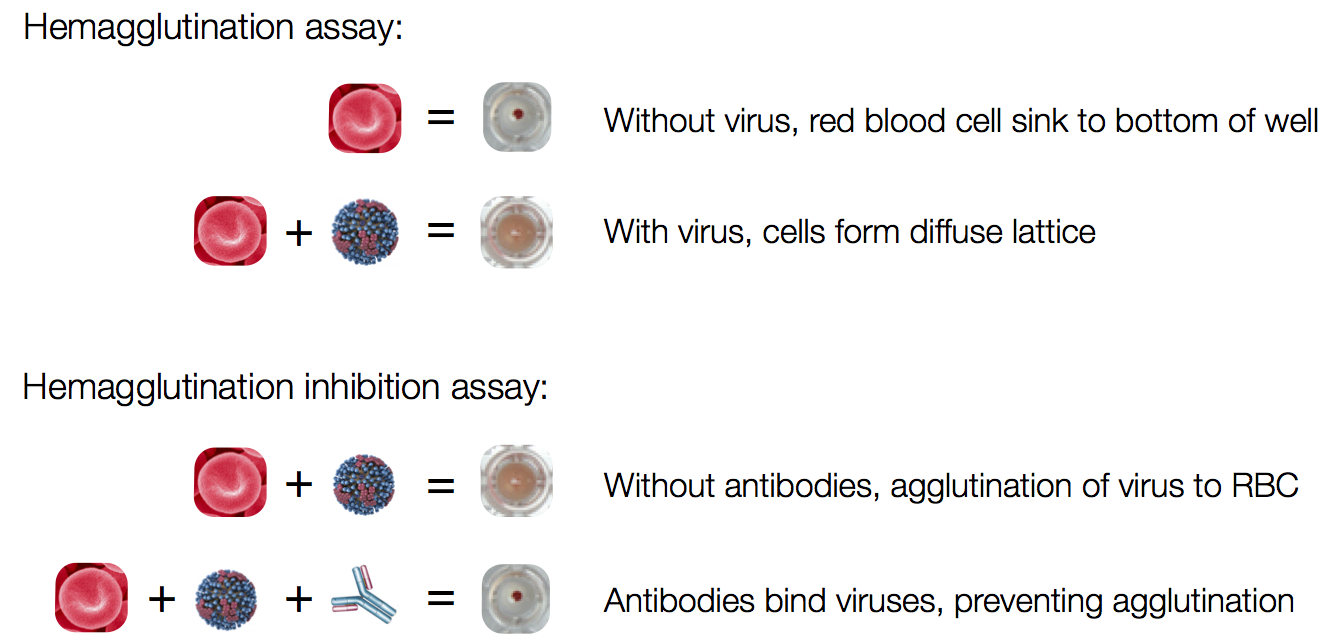
Model is highly predictive of
withheld titer values

Neher et al, PNAS 2016
How does
antigenic evolution
relate to
population dynamics?

Population susceptibility:


Previously circulating:





Population
immunity:













Clade growth:


How does antigenic evolution relate to population dynamics?
Predict clade growth
based on antigenic fitness
Lukzsa and Lassig, Nature, 2014
Growth rate @
next year

Ensemble model of
antigenic fitness
John Huddleston
Antigenic drift
Epitope mutations
Clade expansion
Protein function
+
+
+
Prospective trajectories from
ensemble model of antigenic fitness

John Huddleston
Clade growth rates are well-predicted
John Huddleston

Clade growth vs decline is also well-predicted
John Huddleston

Unlikely to supplant expert predictions, but useful for vaccine candidate selection
So... how well does this generalize?
Contrasting evolutionary dynamics

Dengue (serotype 2)

Influenza (H3N2)
Four (uniform?) serotypes of dengue


Each serotype is genetically diverse
Sanofi vaccine trial efficacy
varies by genotype

Juraska et al, PNAS 2018






Lifelong protection
Temporary
cross-protection
Initial
response
Original antigenic sin drives
dengue case outcomes
After
1-3 yrs



Increased risk
DENV antigenic relationships
are poorly understood
Serotypes are genetically distinct
Clades are genetically distinct
Serotypes are antigenically distinct
Are clades antigenically distinct?
≠
?




≠


≠


How does
dengue evolve
antigenically?
Measuring antigenic relationships


Neutralization

+






+

Replication
Neutralizing antibodies block virus replication
Titers measure antigenic distance

Antigenic similarity = how much sera is required to neutralize viral replication?
Serum



Test viruses






1x
2x
4x
(relative to autologous)
X
X
X
X
X
X



--
Log2
0
1
2
Titer
Titers approximate pairwise
antigenic distance






Experimentally measure how well sera inhibits viral plaque formation



Sera produced by 1st infection
Test viruses
Dengue titers are confusing
Non-human primate sera; 3 months post primary infection
Katzelnick et al, Science 2015

Antigenic distance
(less cross immunity)
Dengue serotypes look antigenically diverse

NHP sera; 3 months post primary infection
= 2-fold change in PRNT50
Katzelnick et al, Science 2015
Where does this variation come from? Does it matter?
Neher et al, PNAS 2016






| 0 | 1 | 2 |
| 1 | 0 | 2 |
| 2 | 2 | 0 |

Titer
between
&





+ avidity + potency
1.5
0.5
Substitution-based model of
antigenic evolution
MRCVGIGSRDFVE...
MRCVGVGNRDHVE...
Model is highly predictive of
withheld titer values
Antigenic mutations occur between and within serotypes

Each serotype of dengue contains multiple
distinct antigenic phenotypes

How does
antigenic diversity
impact dengue
population dynamics?
Serotypes cycle through populations
Bell et al., bioRxiv 2018

Does antigenic fitness
predict serotype growth & decline?
Lukzsa and Lassig, Nature 2014
Growth rate @
next 5 years
How antigenically distant
is from what has recently circulated?


Antigenic novelty drives
viral fitness and serotype turnover
Bell et al., bioRxiv 2018


Dengue
Flu (H3N2)
12 years
500 years
4 titer units*
11 titer units
* likely underestimated
Dengue antigenic evolution is ongoing but slow

Methods summary & further reading:
Antigenic cartography
Visualize antigenic space via a low-dimensional embedding
Evolution-based models
Assign antigenic change to branches or substitions, interpolate across dataset
Population dynamics
Antigenic fitness & clade turnover
Acknowledgements






Slides: The Noun Project & RevealJS
Influenza: Trevor Bedford, Richard Neher, John Huddleston
Dengue: Trevor Bedford, Leah Katzelnick, Gytis Dudas, James Hadfield, John Huddleston, Alli Black, Barney Potter, Louise Moncla, Richard Neher
CZI comp bio: Jeremy Freeman, Deep Ganguli, Nick Sofroniev, Genevieve Haliburton, Ambrose Carr
Tools for Open Science
Or: the tools I wish I knew about when I started


Gitkraken: to make git less confusing

Protocols.io: version, share,
collaborate, cite & adapt protocols


Figshare: version, cite & share datasets

ARTICnetwork: resources for
genomic epidemiology

Nextstrain: interactive, shareable explorer for pathogen genomic data



Rxivist: finding awesome preprints
Biorender & Noun Project:
better visual communication


Questions?
@sidneymbell



phylo-workshop-san-diego-2019
By Sidney Bell
phylo-workshop-san-diego-2019
- 1,863



