From structure to function: leveraging embedded multimodal imaging to explore brain connectivity

| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
AIRMM 2025, Lecco, 26.03.2025


Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, The Netherlands; Open Science Special Interest Group (OHBM); physiopy (https://github.com/physiopy)




I have nothing to disclose.
This is a new chapter



This is a new chapter



Take home #0
This is a take home message
Connectivity across modalities



BOLD signal: "Functional Networks"
Biswal et al., 1995 (Magn. Reson. Med.)
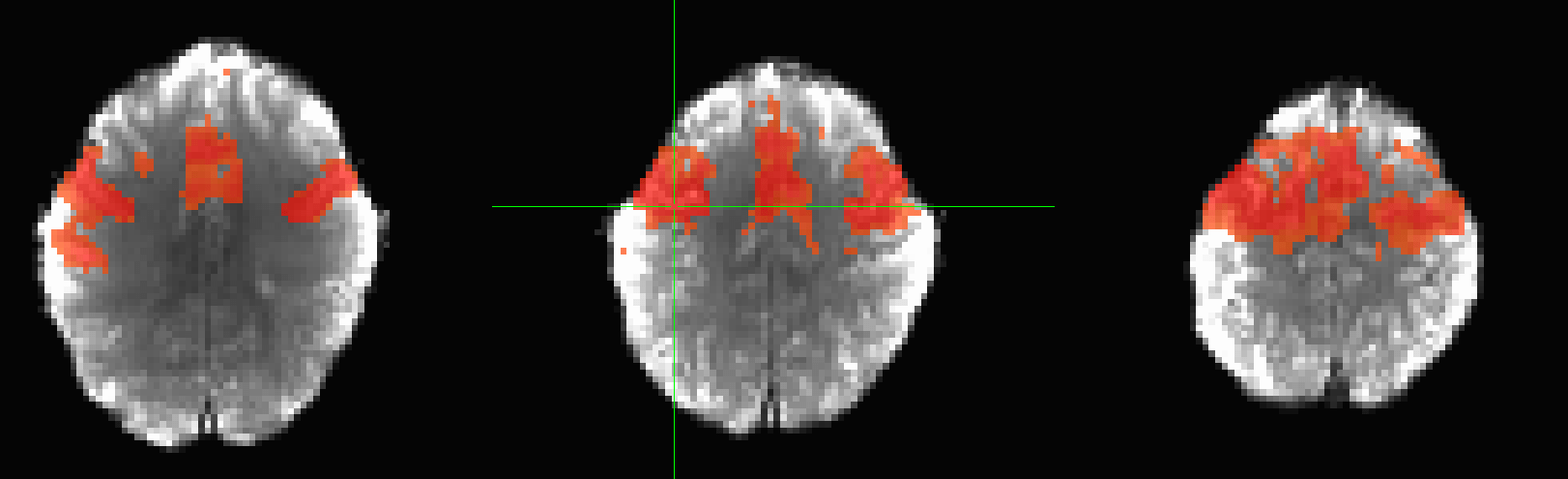
Pearson correlation:



Whole brain functional connectivity
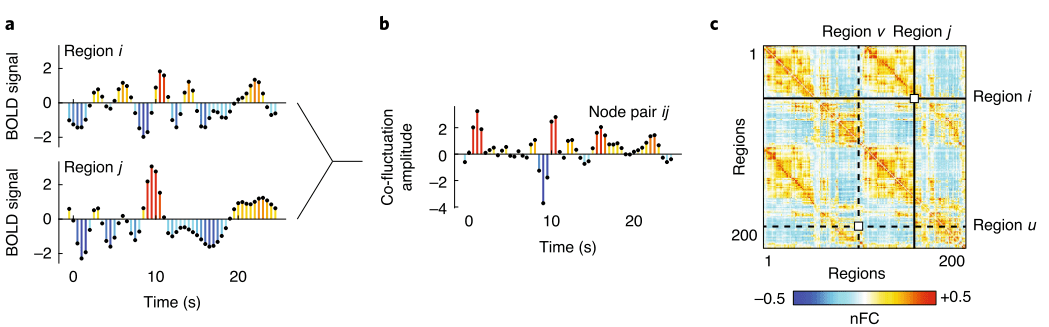







Images adapted from Moghimi et al., 2022 (J. Neurophysiol.) and Faskowitz et al., 2020 (Nat. Neurosci.)
Whole brain structural connectivity
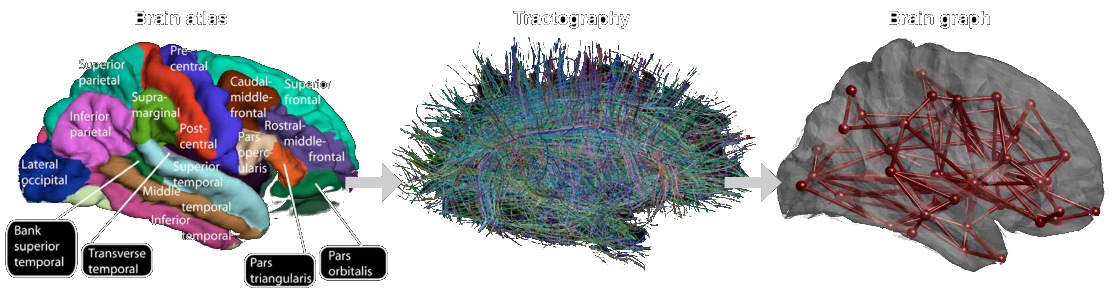
Huang et al., 2018 (Proc. IEEE)





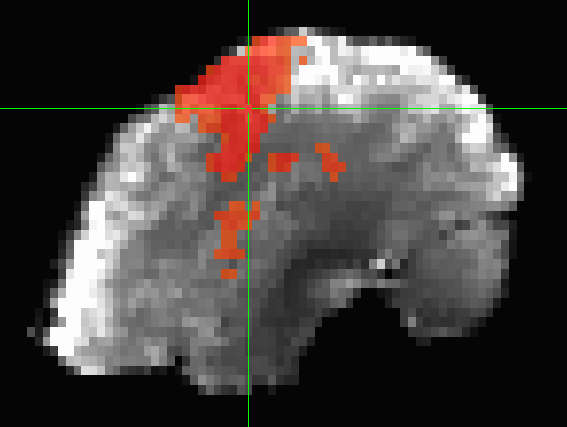
(i)EEG connectivity



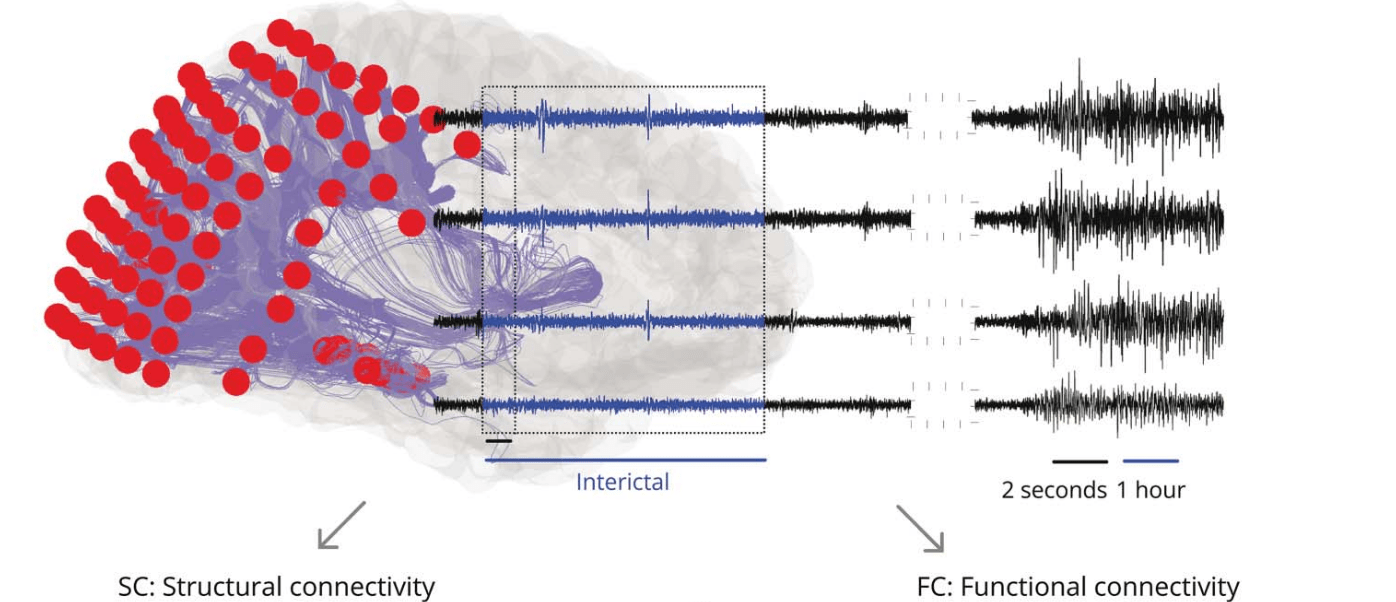
Images adapted from Sinha et al., 2023 (Neurol.)
(i)EEG connectivity




Images adapted from Sinha et al., 2023 (Neurol.)
Comparing Structural and Functional Connectivity



Baum et al., 2020 (Proc. Natl. Acad. Sci.)
Easy-peasy Structure-Function Coupling (SFC)
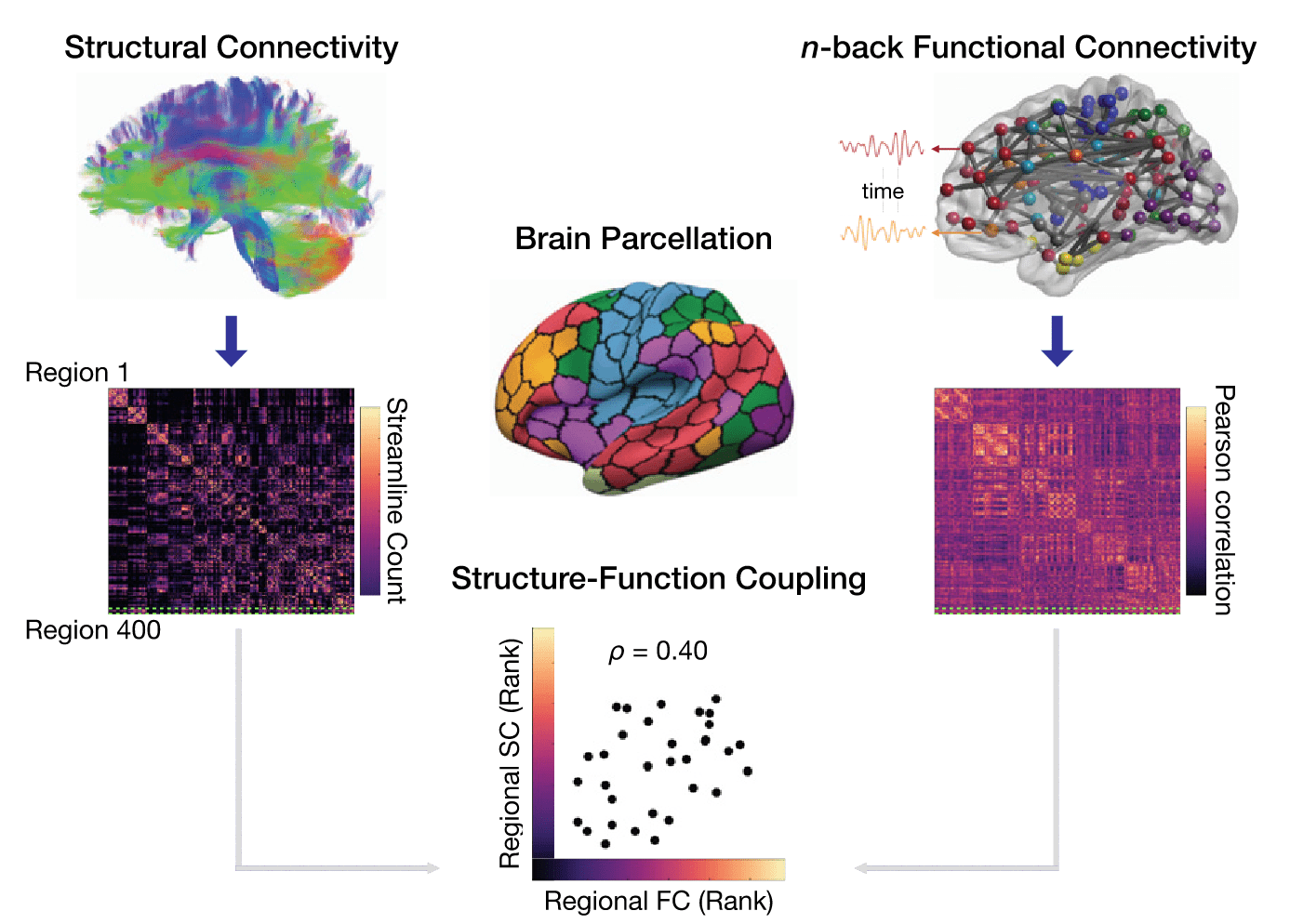



Baum et al., 2020 (Proc. Natl. Acad. Sci.)
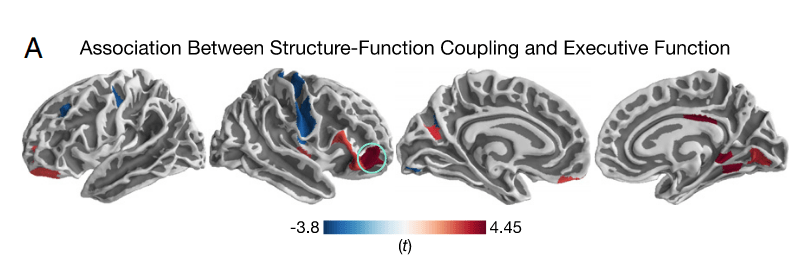
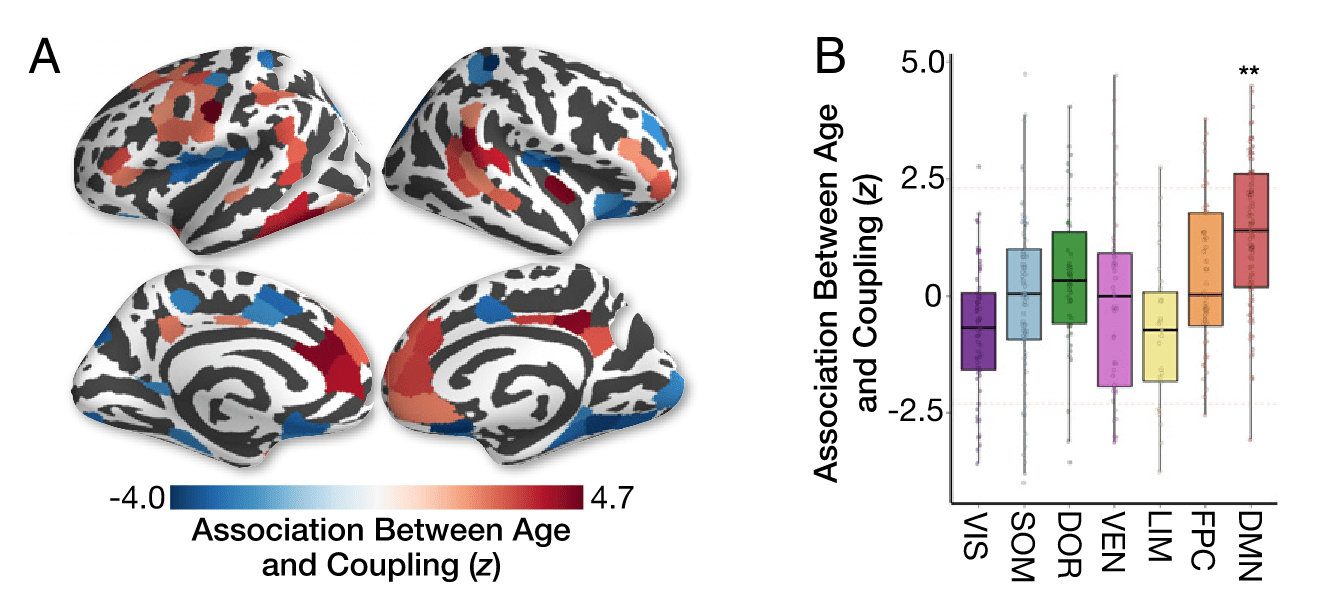
SFC over the life span



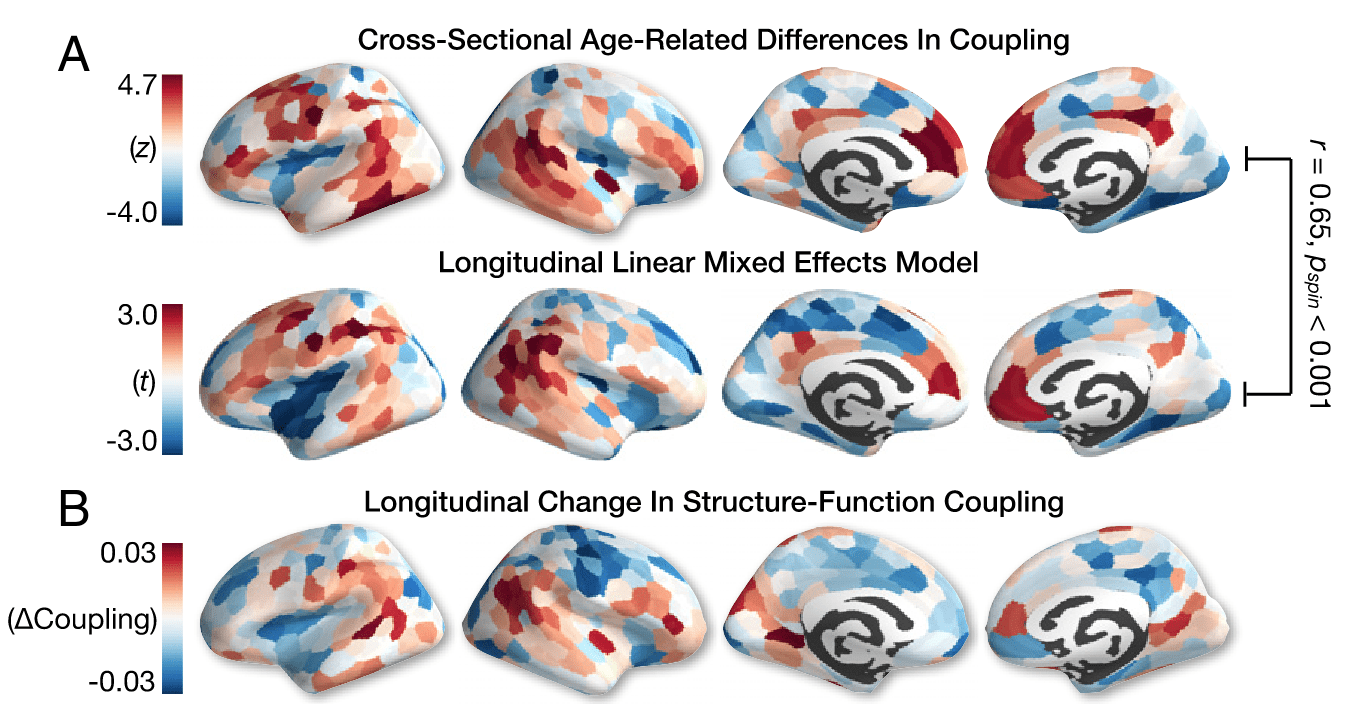
727 participants (420 F), 8 to 23 y.o., BOLD FC & dMRI
Sinha et al., 2023 (Neurol.)
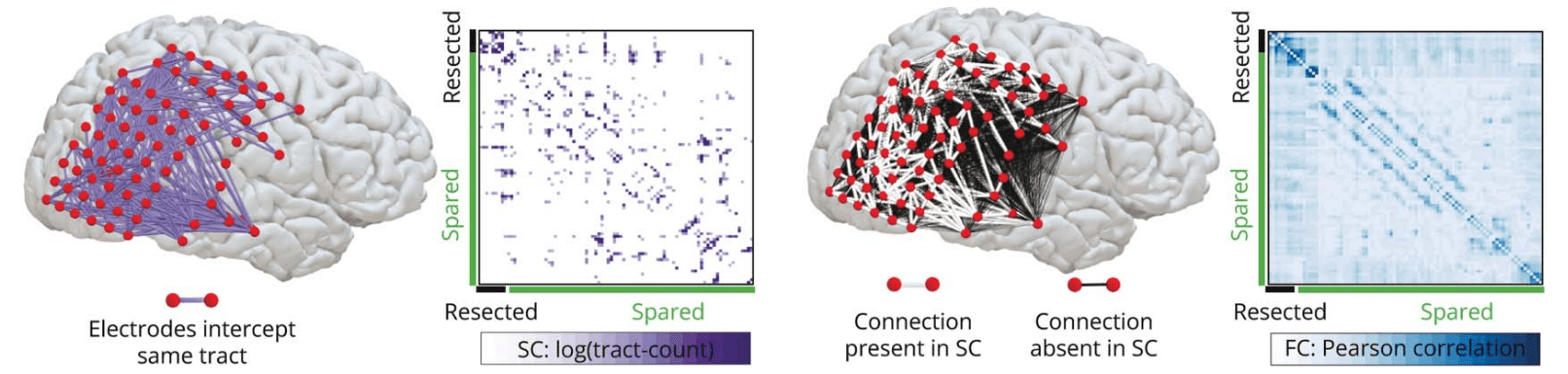
SFC and post-surgery seizure outcome



15 SF (7F) 24 nSF (16)
iEEG-FC (1h) & dMRI

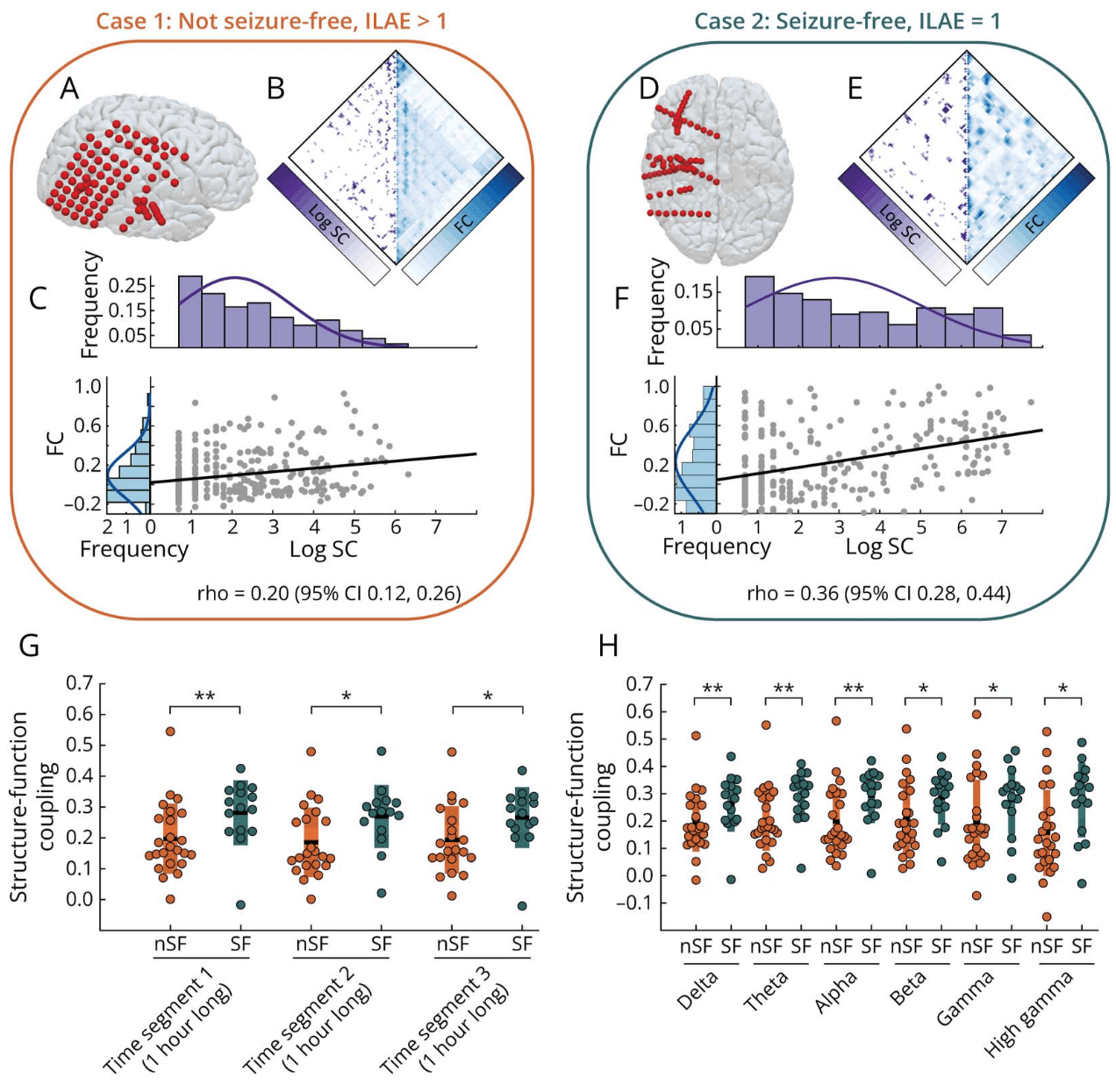
SF = Seizure Free nSF = non-Seizure Free
Structure-function coupling in EEG/dMRI




Caveat
Images adapted from Sinha et al., 2023 (Neurol.)


Sparse
\( [0,+∞) \)
Highly skewed distribution
Non-Euclidean
Dense
\( [-1,+1] \)
Somewhat normal distribution
Euclidean



Take home #1
Observing Multimodal Connectivity can be
as easy as computing a correlation.



HOWEVER,
there are a few caveats you should be aware of when doing so.
Graph Signal Processing



From timeseries to graphs
x1
x2
x3
x8
...



From timeseries to graphs
x1
x2
x3
x8
...



From timeseries to graphs
x1
x2
x3
x8
...



From timeseries to graphs
x1
x2
x3
x8
...
x1
x2
x3
x8
...
x1
x2
x3
x8
...



From timeseries to graphs
x1
x2
x3
x8
...
x1
x2
x3
x8
...
x1
x2
x3
...
x8



From timeseries to graphs
x1
x2
x3
x8
...
x1
x2
x3
x8
...
x1
x2
x3
...
x8



From timeseries to graphs
x1
x2
x3
x8
...
x1
x2
x3
x8
...
x1
x2
x3
...
x8



From timeseries to graphs
x1
x2
x3
x8
...
x1
x2
x3
x8
...
x1
x2
x3
...
x8



From timeseries to graphs
x1
x2
x3
x8
...
x1
x2
x3
x8
...
x1
x2
x3
...
x8



Graph Signal Processing
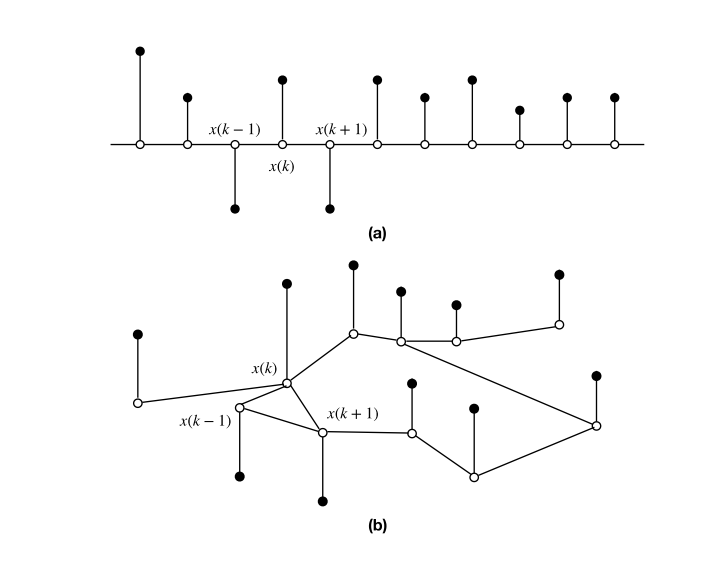
Ortega 2022 (Introduction to Graph signal Processing)



White dots = nodes
Black dots = timeseries value
Embedding brain function
into the brain structure



Image courtesy of Maria Giulia Preti; Preti et al., 2019 (Nat. Commun.)
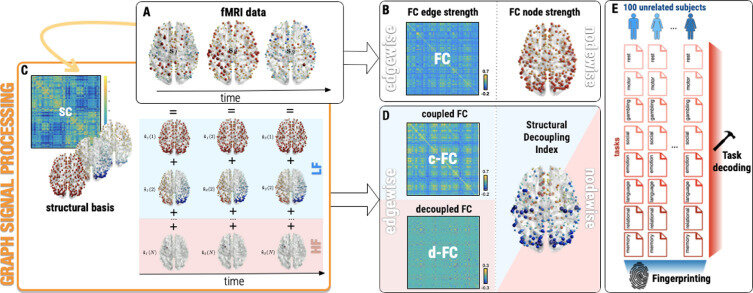
Graph Signal Processing: Decomposition
Decomposition
to access
"graph" spectral domain
(e.g. Principal Components)





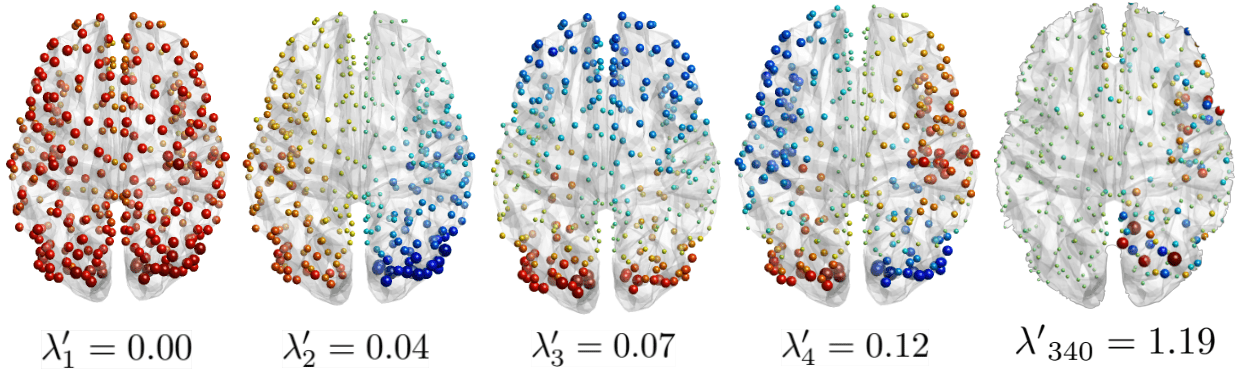
Huang et al., 2018 (Proc. IEEE)
Graph Signal Processing: Filtering
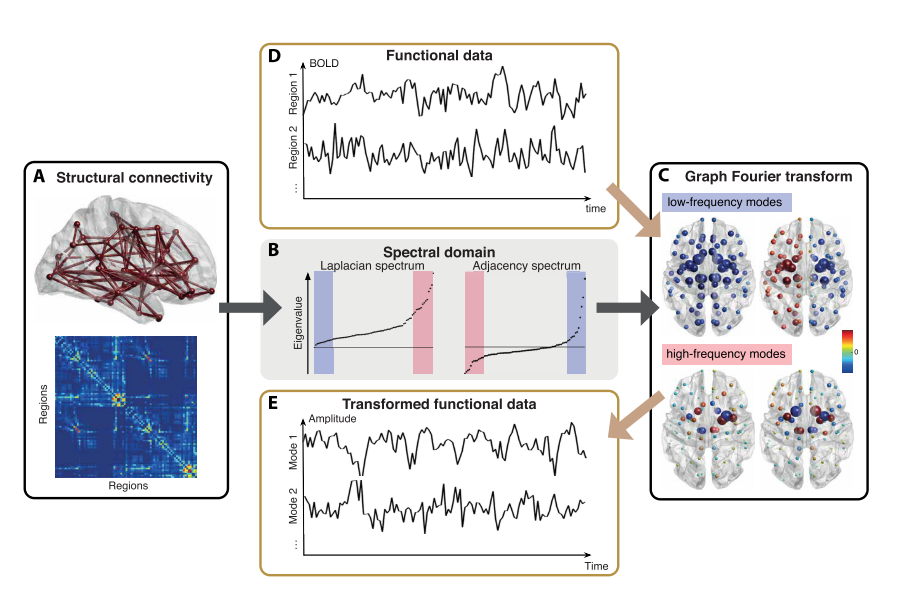



Preti et al., 2019 (Nat. Commun.)
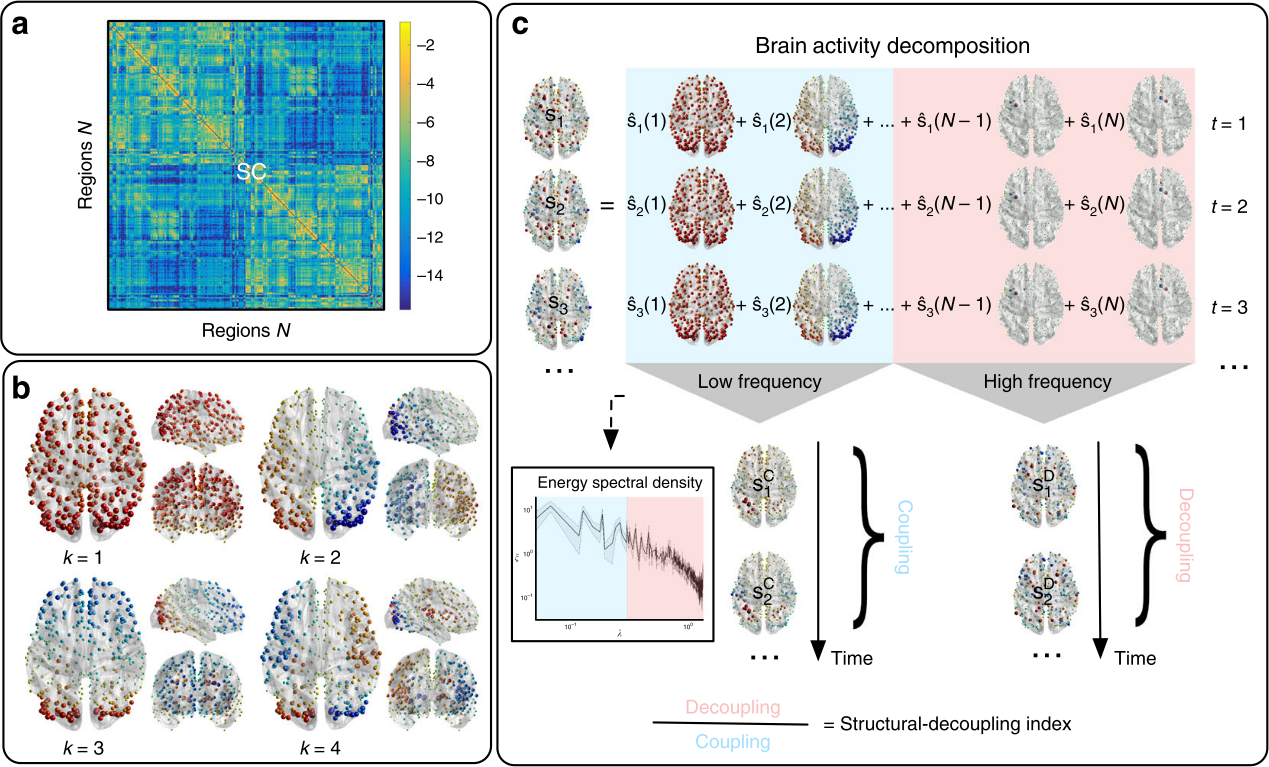
Graph Signal Processing: Structural decoupling index (SDI)



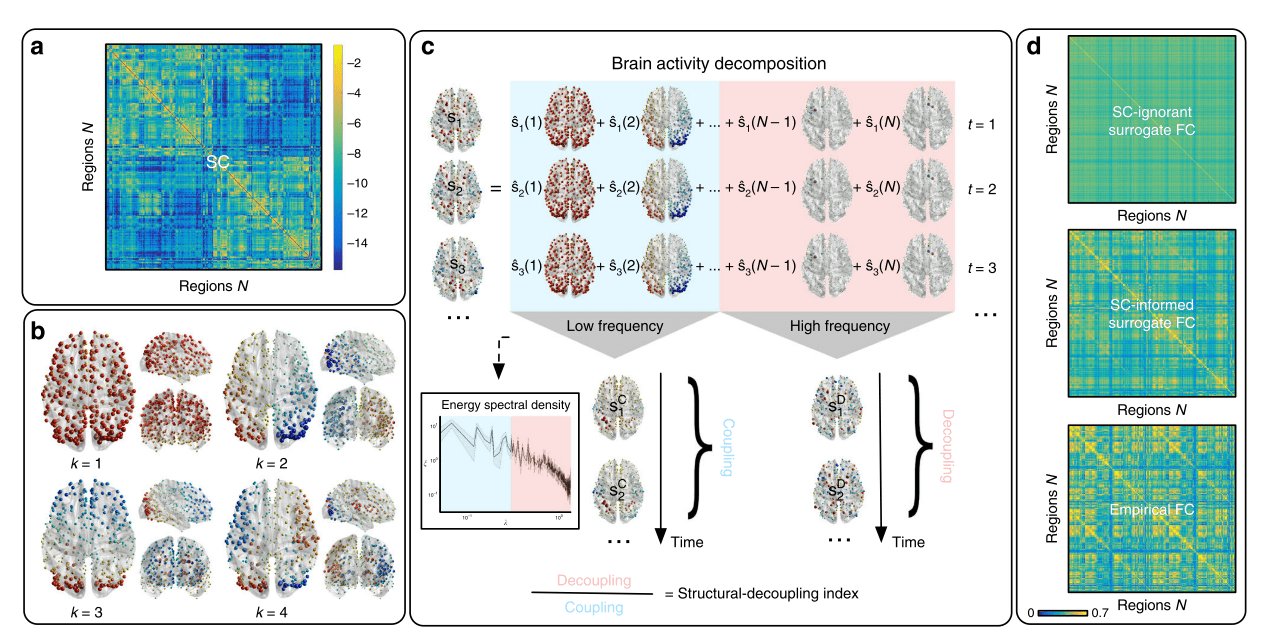
1. Cioli et al., 2014 (PLoS One), Mesmoudi et al. 2013 (PLoS One); Preti et al., 2019 (Nat. Commun.)
Graph Signal Processing: Structural decoupling index (SDI)



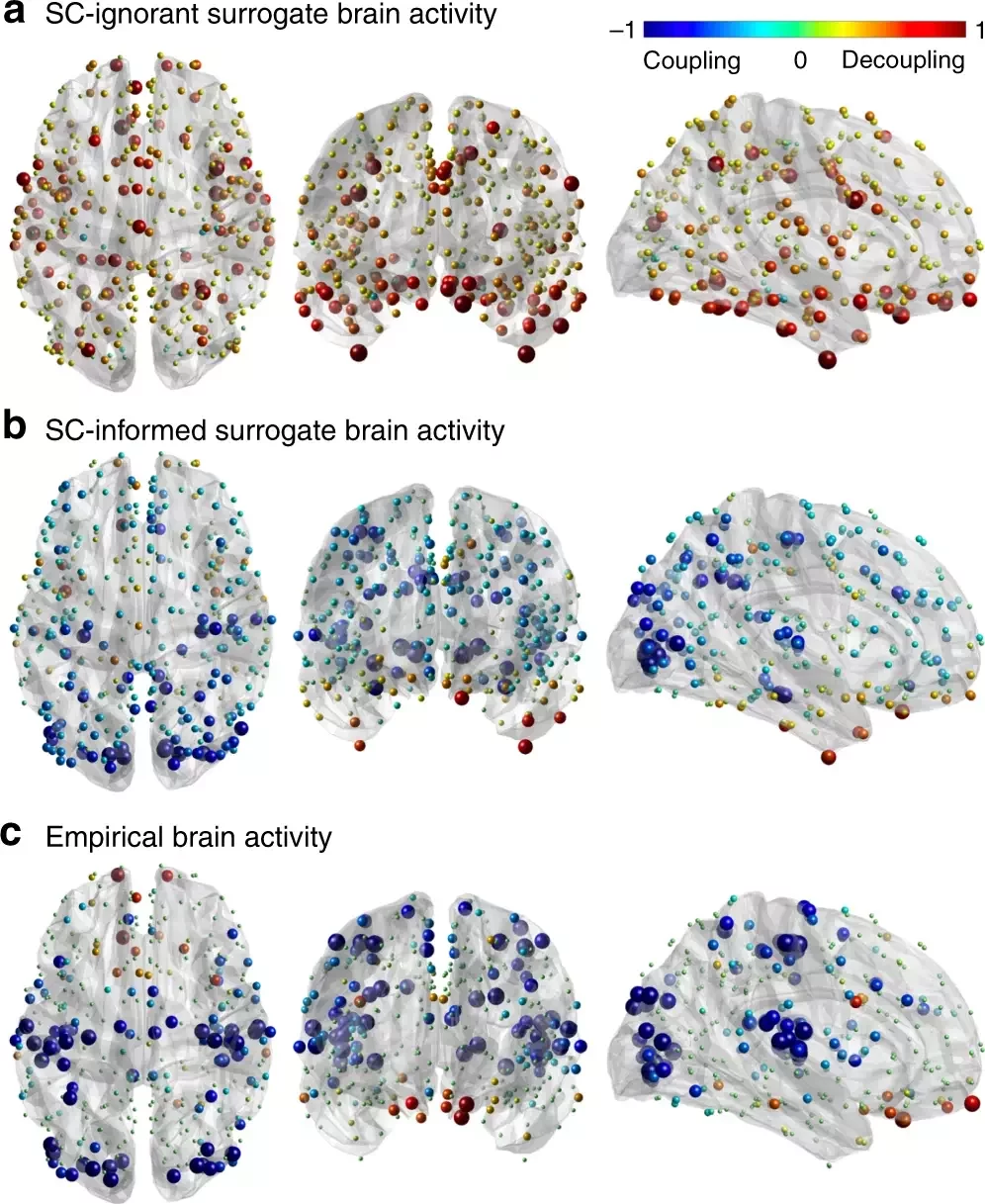
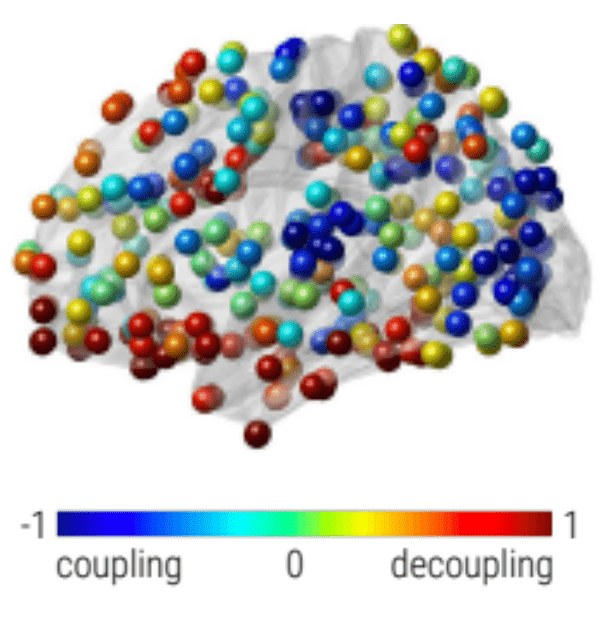
Gene expression¹
SDI²
Rapid responses Slow (sustained) responses
High functions Low functions
Cioli et al., 2014 (PLoS One); Preti et al., 2019 (Nat. Commun.)
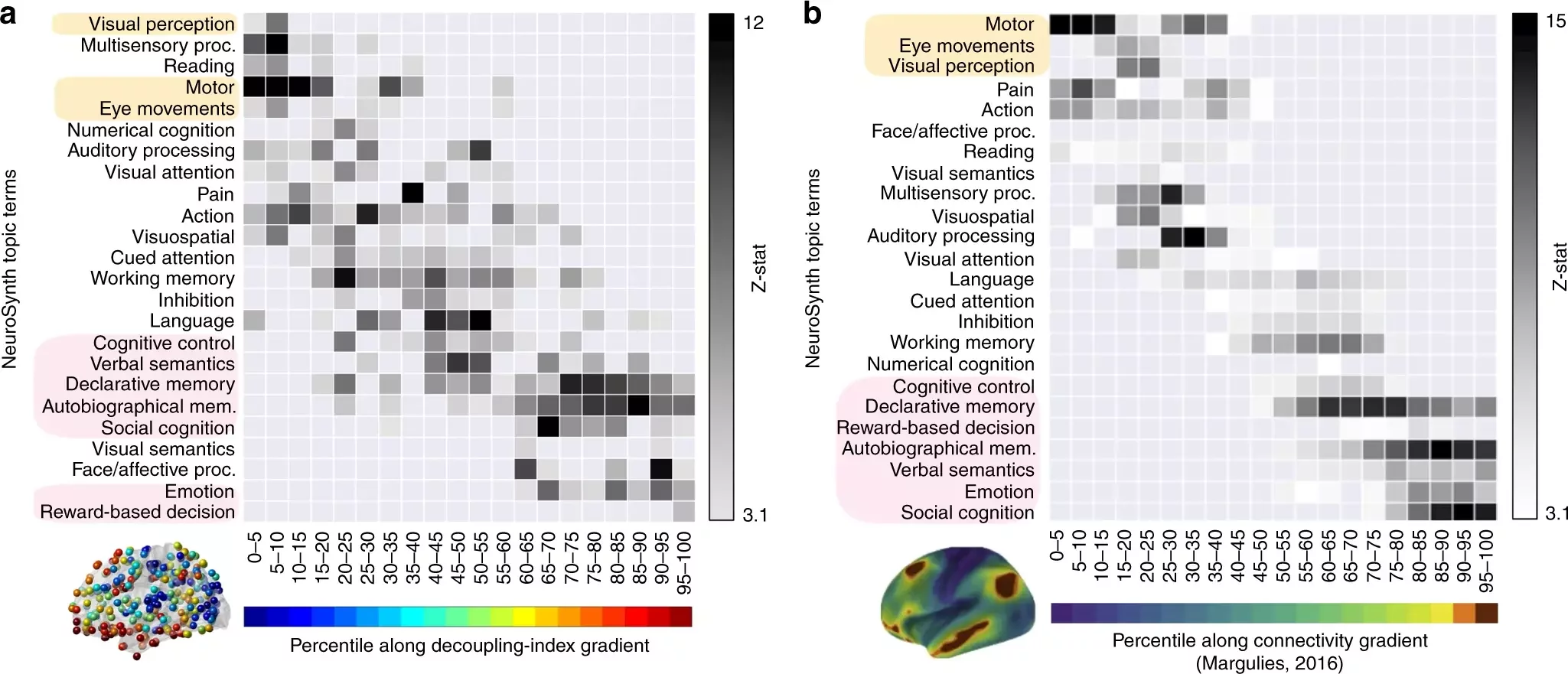
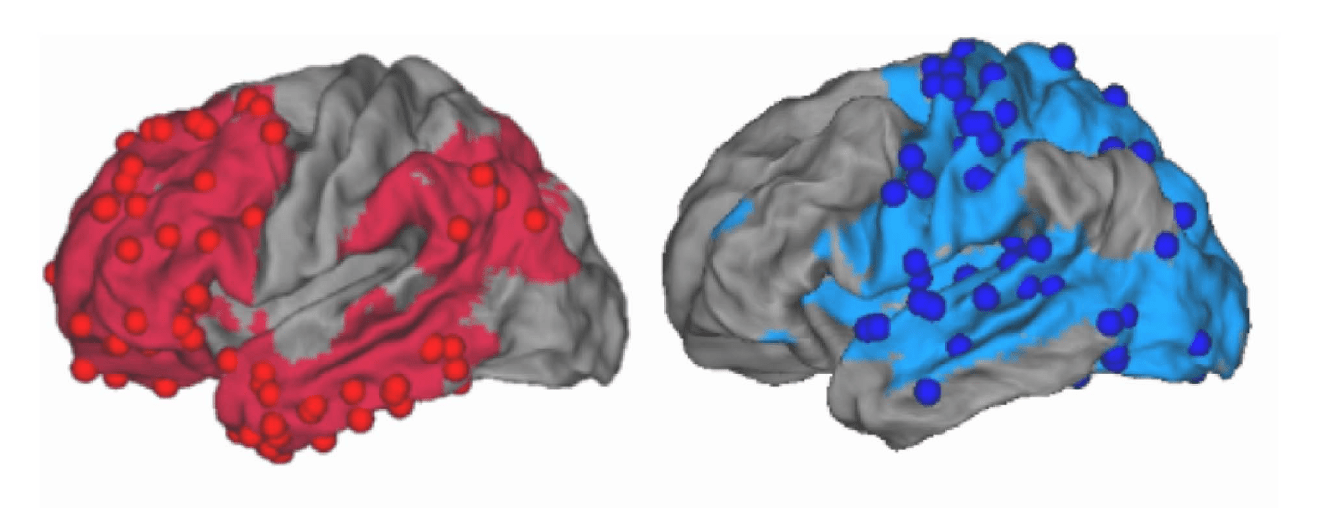
Graph Signal Processing: Subject-Task Identification



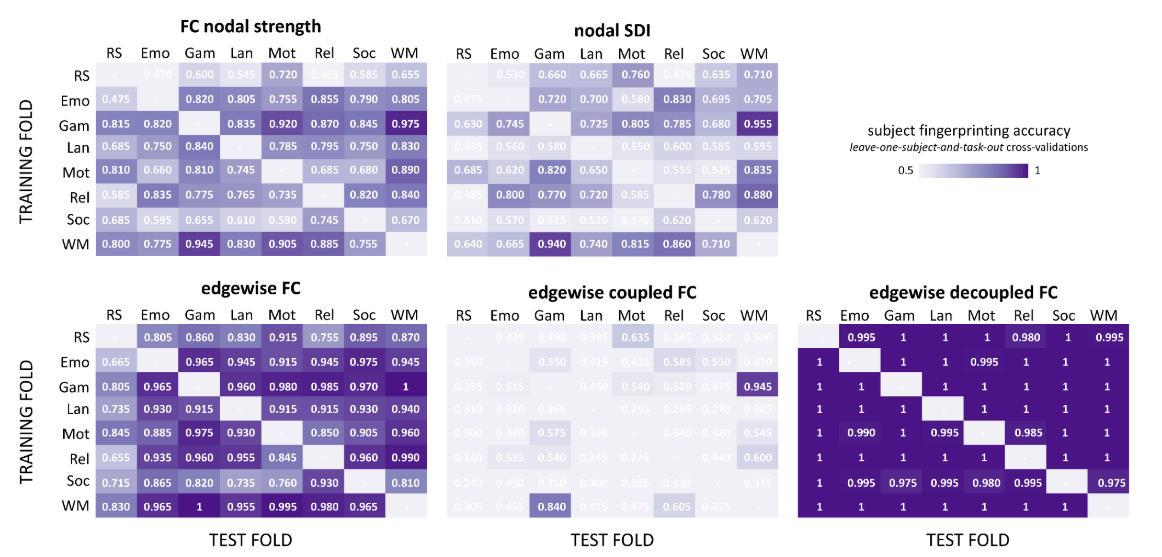
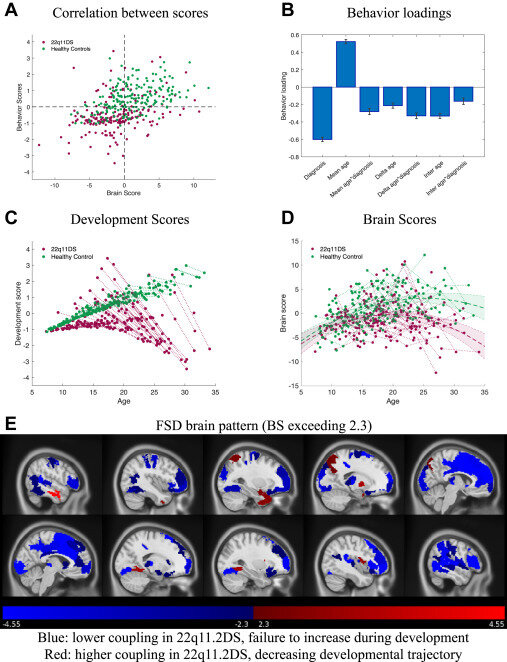
SDI in 22q11.2 deletion syndrome
Forrer et al., 2024 (Biol. Psychiatry)



233 subjects (118F), 108 22q11.2DS (50F), 7-34 y.o.
BOLD fMRI & DWI
SDI in Parkinson




Time-resolved SDI in movies and rest¹
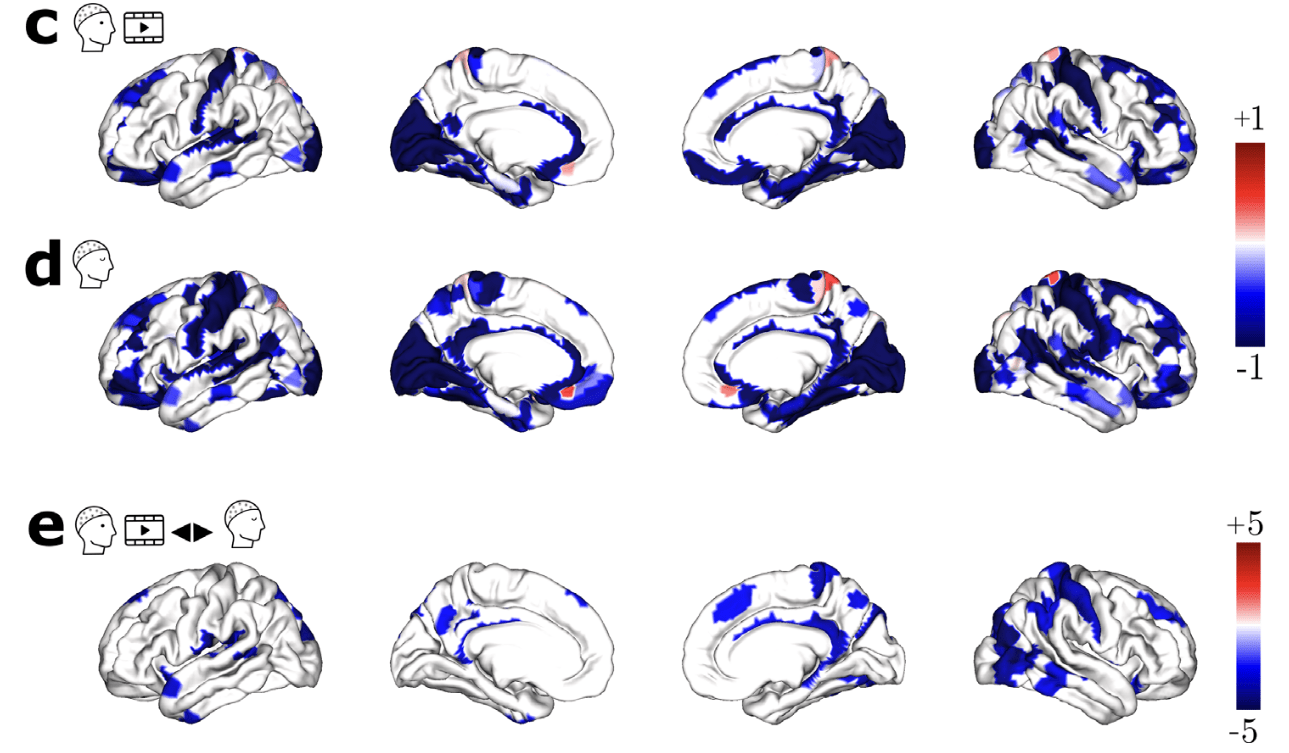
1. Subramani et al., 2025 (Imaging Neurosci.); 2. Langer et al., 2017 (Sci. Data)
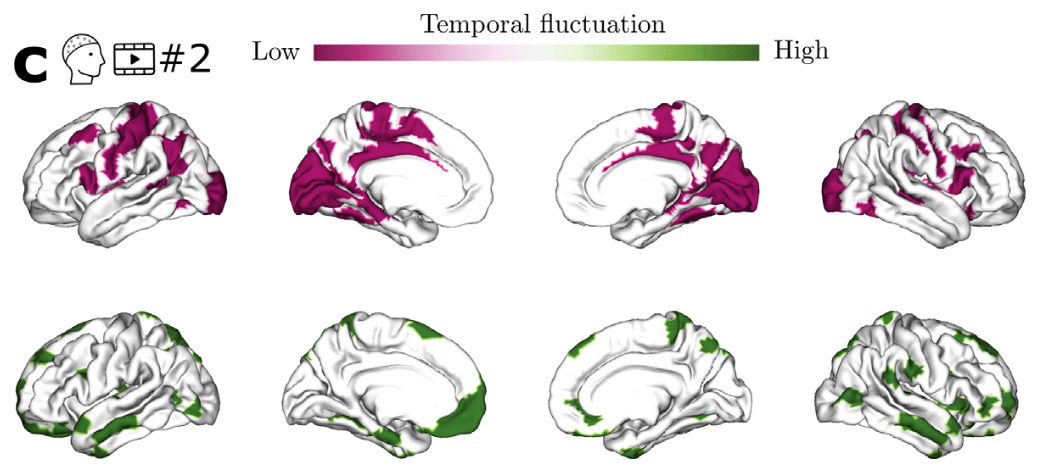



Healthy Brain
Network² data
EEG & DWI
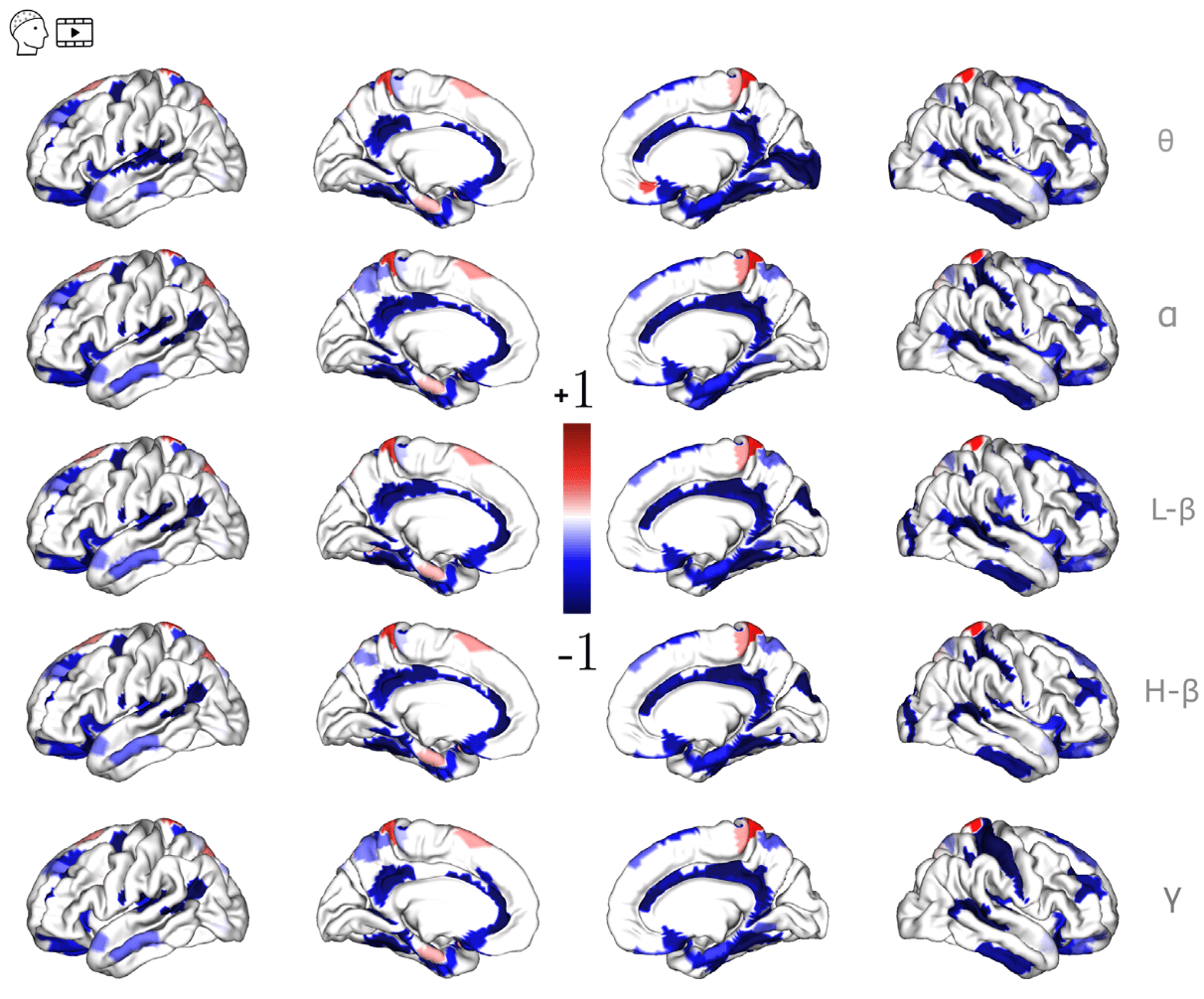
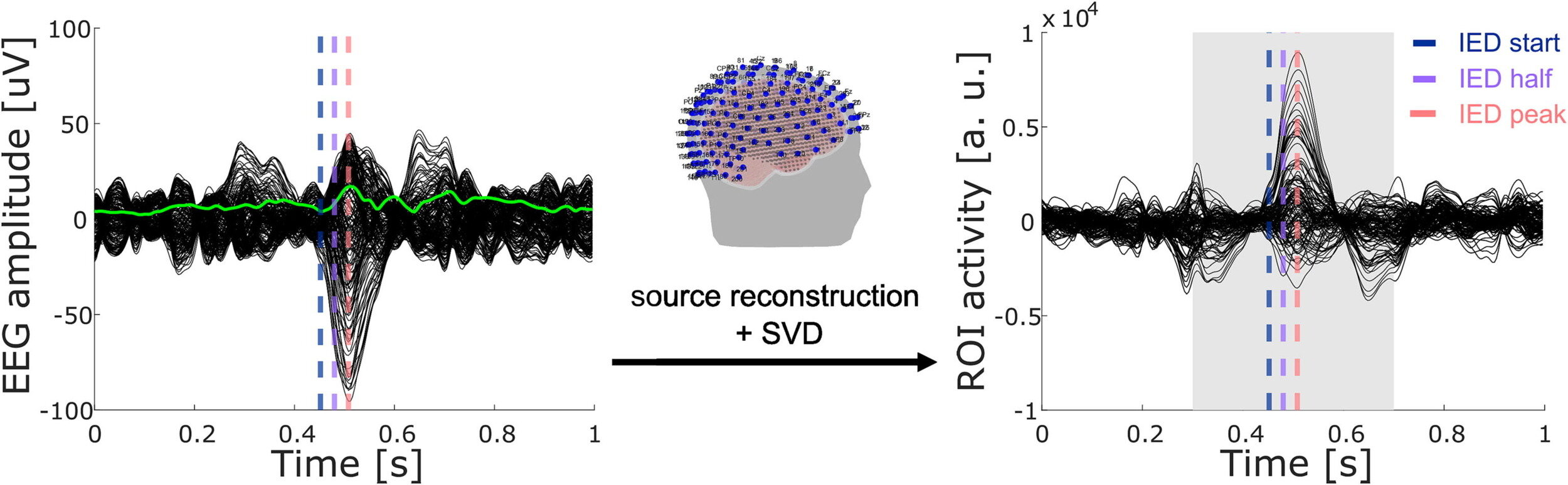
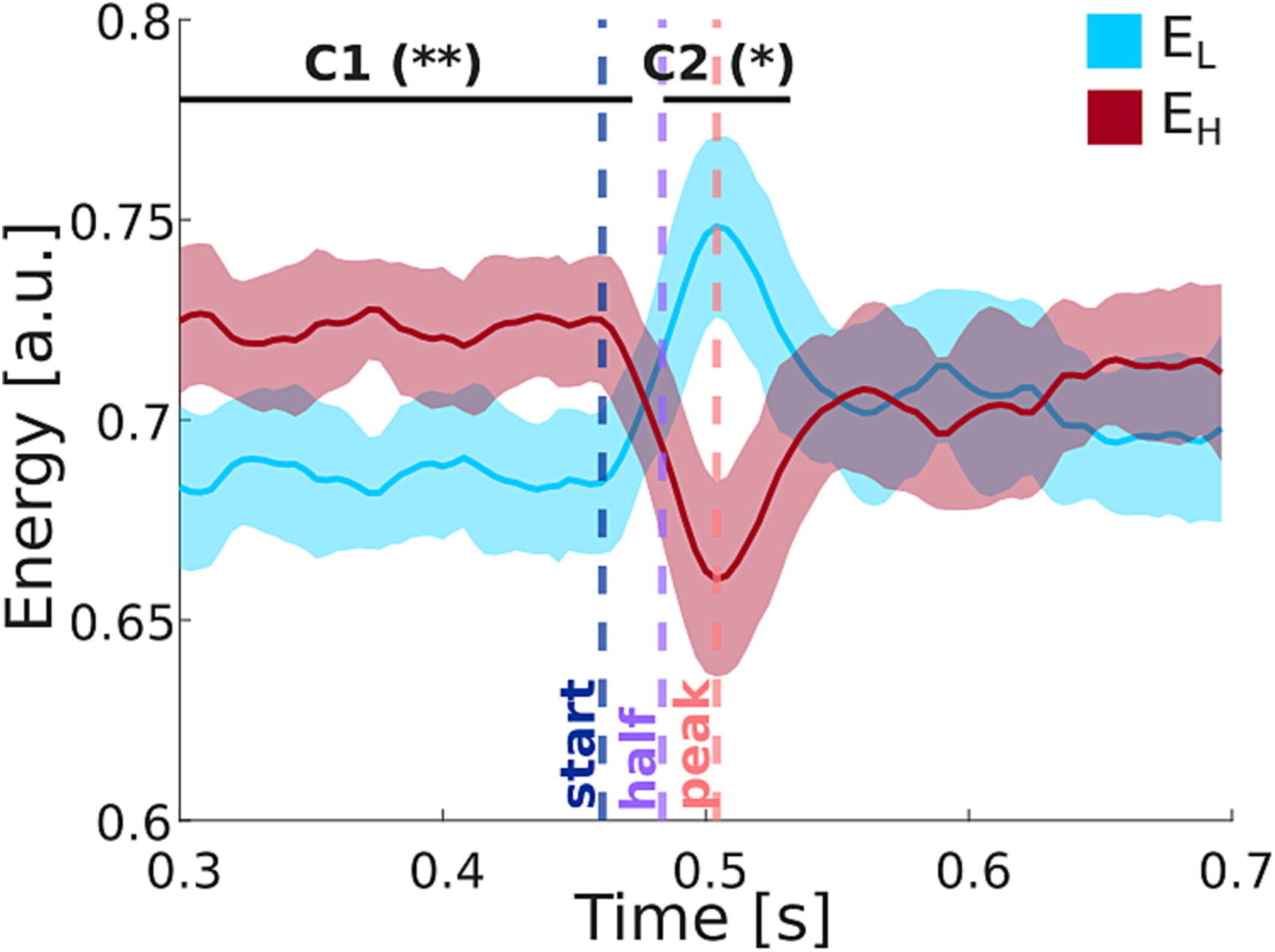
SDI in temporal lobe epilepsy (TLE)
Rigoni et al., 2023 (Clin. Neurophysiol.)



17 patients (5F),
8 left TLE,
15–56 y.o.
EEG & DWI
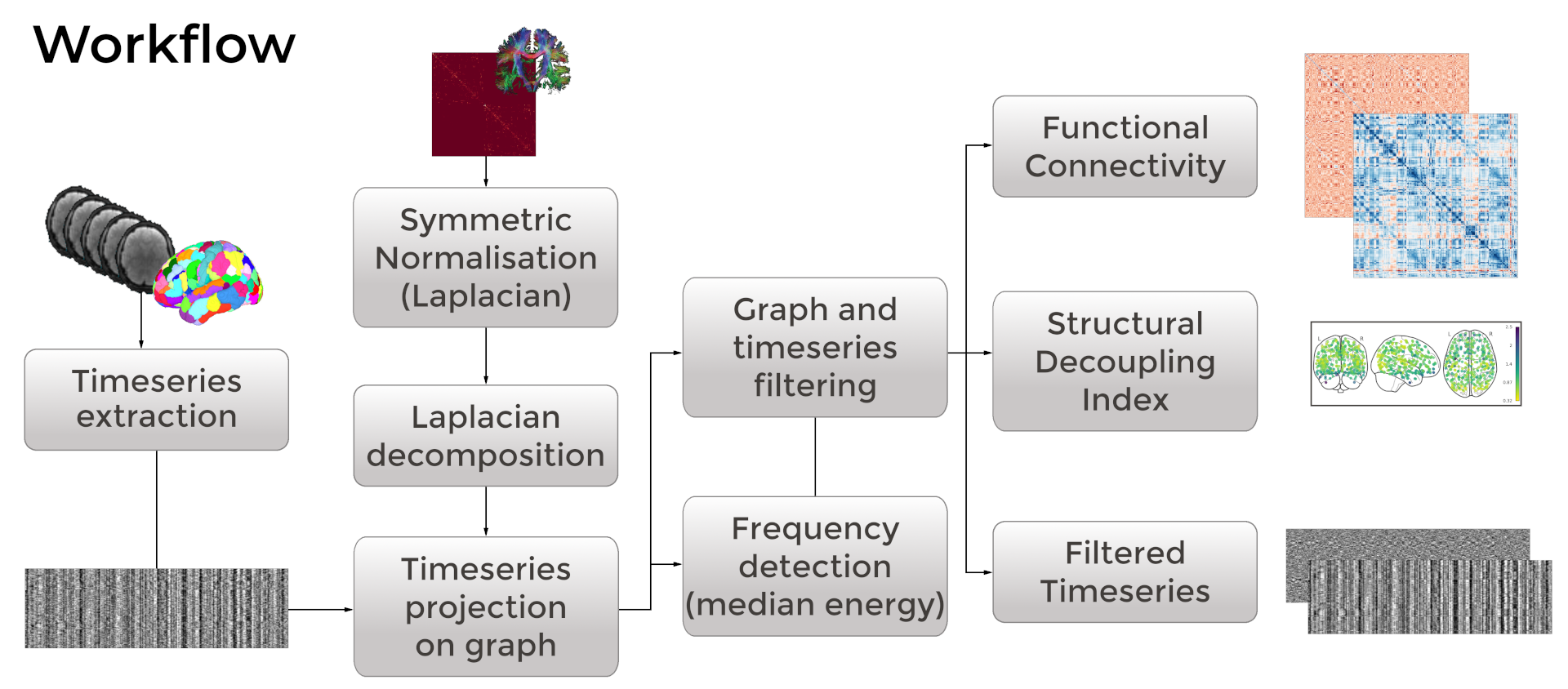
pip install nigsp[all]
nigsp -f timeseries.nii.gz -s sc_mtx.tsv \
-a atlas.nii.gz --informed-surrogates -n 1000Moia et al. (2023) Zenodo; based on Preti, Van De Ville (2019) Nat. Commun. and Griffa et al. (2022) Neuroimage
Applying Graph Signal Processing: NiGSP



Take home #2
We can leverage Graph Signal Processing
to better explore more complex spatio-temporal features of timeseries embedded in graphs.*



BOLD fMRI
M/EEG
fNIRS
Tractography
EEG caps
Anatomy
*And it's still as easy as installing a python package
Thanks to...
That's all folks!

| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |


...you for the (sustained) attention!
...the MR-methods group @UM


...the MIP:Lab @ EPFL



Find the presentation at:
slides.com/smoia/
airmm2025/scroll

Any question [/opinions/objections/...]?
Find the presentation at:
slides.com/smoia/
airmm2025/scroll

1. Multimodal Connectivity analysis is as easy as a correlation of two matrices, with important caveats.
2. Graph Signal Processing is a better approach to model multimodal relationships.
Take home messages
Multimodal Imaging across structure and function
By Stefano Moia
Multimodal Imaging across structure and function
CC-BY 4.0 Stefano Moia, 2025. Images are property of the original authors and should be shared following their respective licences. This presentation is otherwise licensed under CC BY 4.0. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/
- 219



