Practical intro to computationally reproducible science
Maastricht, 03.02.2025

| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |


Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, The Netherlands; Open Science Special Interest Group (OHBM); physiopy (https://github.com/physiopy)




This is a new chapter



This is a new chapter



Take home #0
This is a take home message
This is a new chapter



Take home #0
This is a take home message
This is practical!
Disclaimers
1. I have a bias towards the core tenets of Open and Reproducible Science as better scientific practices.
2. Practices are changing (already) and the landscape is vast. This is a small introduction to containers and git, but there are other ways to do these same things.






(?)
1. Reproducibility
Replicable, Robust, Reproducible, Generalisable

The Turing Way Community, & Scriberia, 2022 (Zenodo). Illustrations from The Turing Way (CC-BY 4.0)
Guaranteeing reproducibility is important for "reusable, transparent" research.






We have a problem.
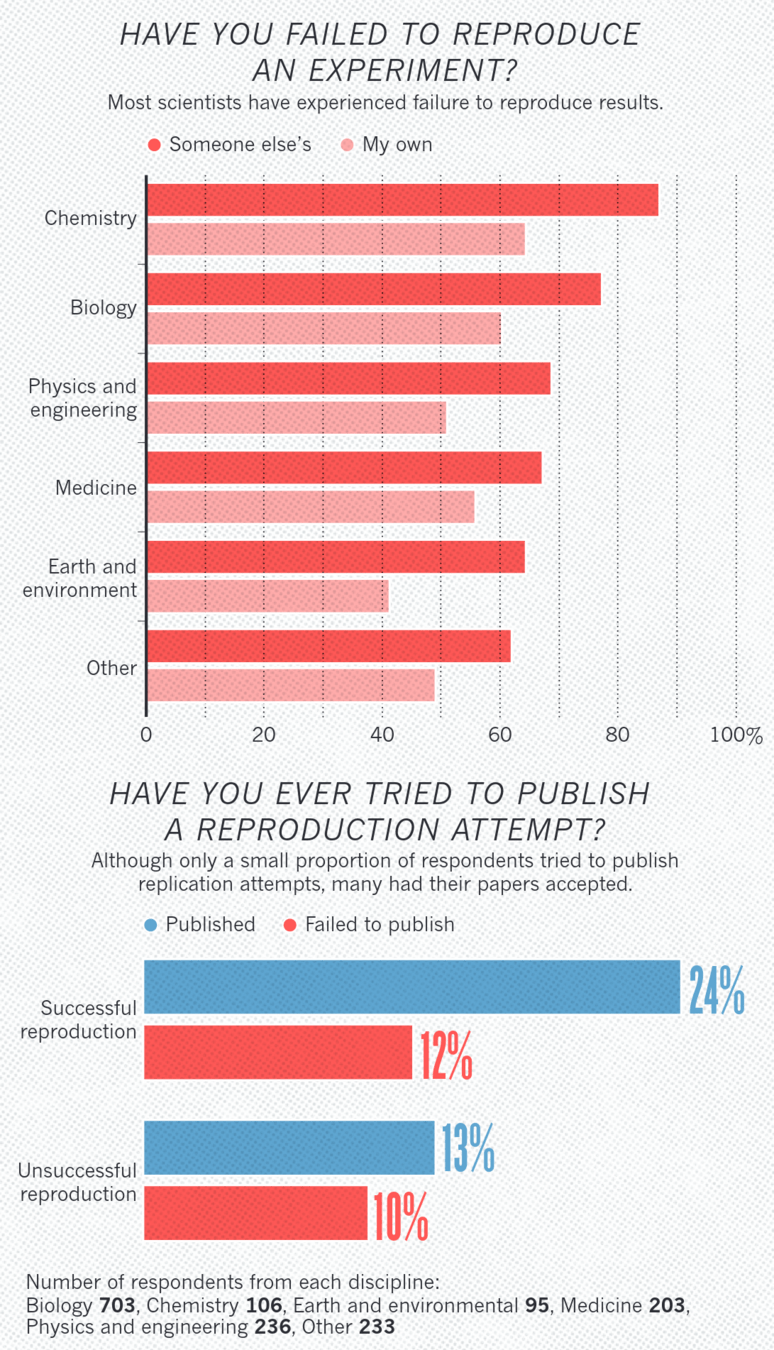
2016: Survey by Nature¹:
- ~70% of researchers failed to reproduce other's results,
- 50%+ failed to reproduce their own
1. Baker 2016 (Nature)



Reproducible?
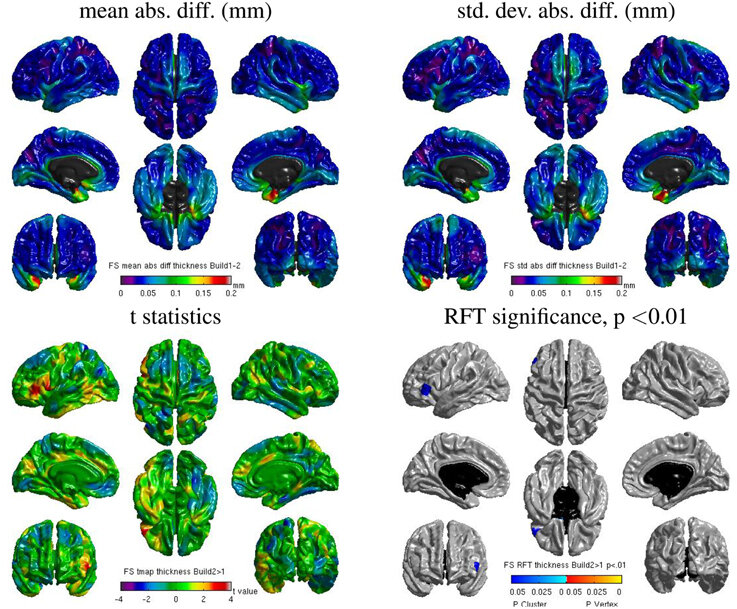
Same hardware, two Freesurfer builds (different glibc version)
Difference in estimated cortical tickness.¹
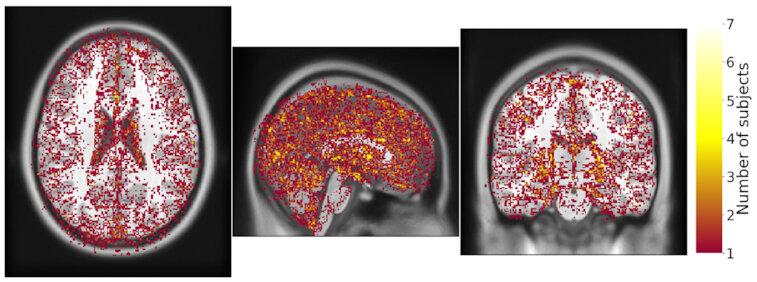
Same hardware, same FSL version, two glibc versions
Difference in estimated tissue segmentation.²
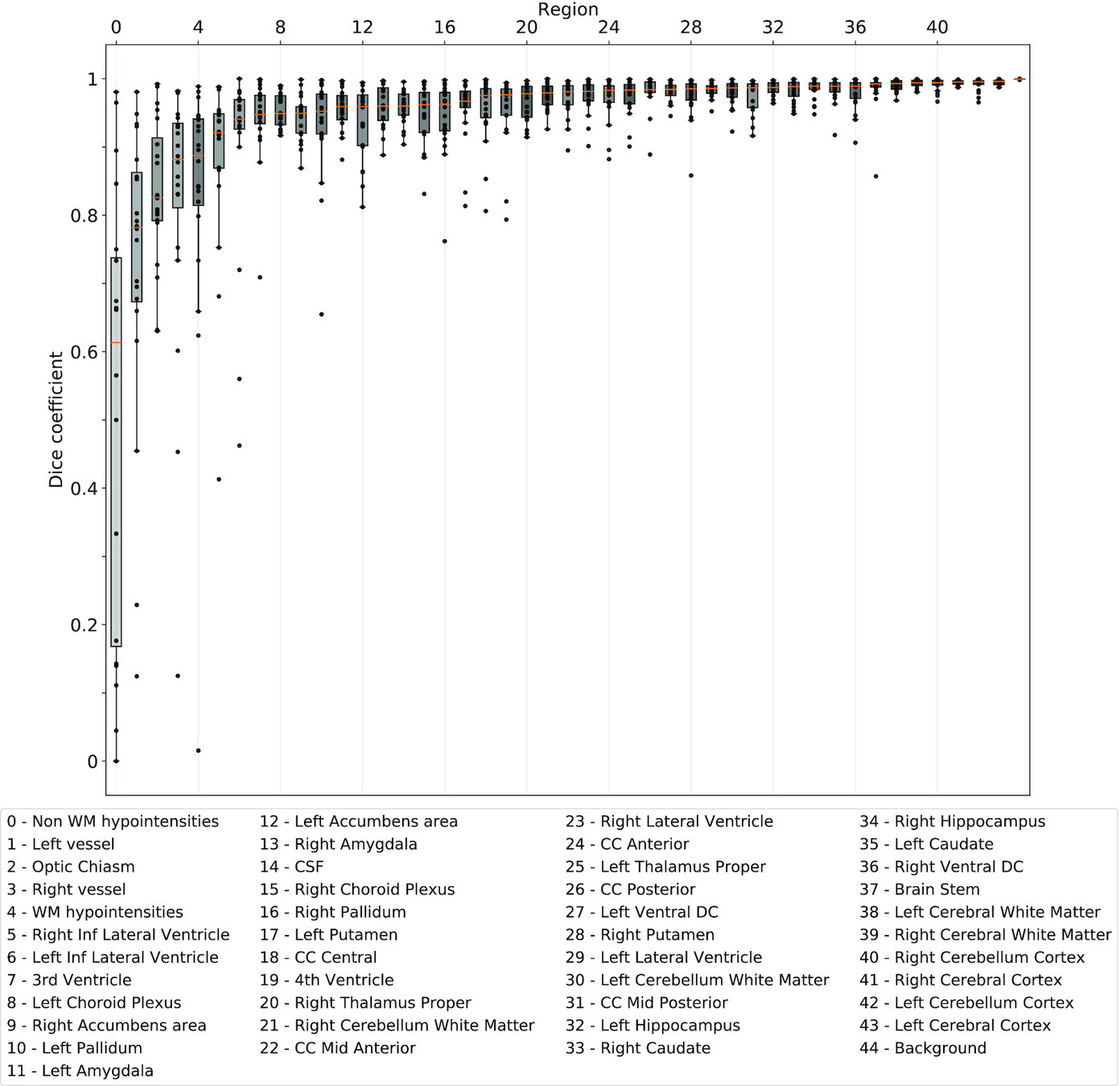
Same hardware, two Freesurfer builds (two glibc versions)
Difference in estimated parcellation.²
1. Glatard, et al., 2015 (Front. Neuroinform.) 2. Ali, et al., 2021 (Gigascience)









(Some) Issues
- Human mistakes / bugs: Objective is not not-human
- Different (algorithmic) implementations: Researchers degrees of freedom
- Different practices in reporting results: Thresholding
- Unavailable data / code / materials / information / procedures
-
Novelty seeking and Publish or perish culture of academia
-
Null results rejection → Bias toward positive results
- Bad "habits": P-hacking, data dredging, data fishing, HARKing, ..., fraud



Take home #1
Don't think that because your analysis "works", it's reproducible.
Make it reproducible.



2. Containers



Containerisation (vs VM)


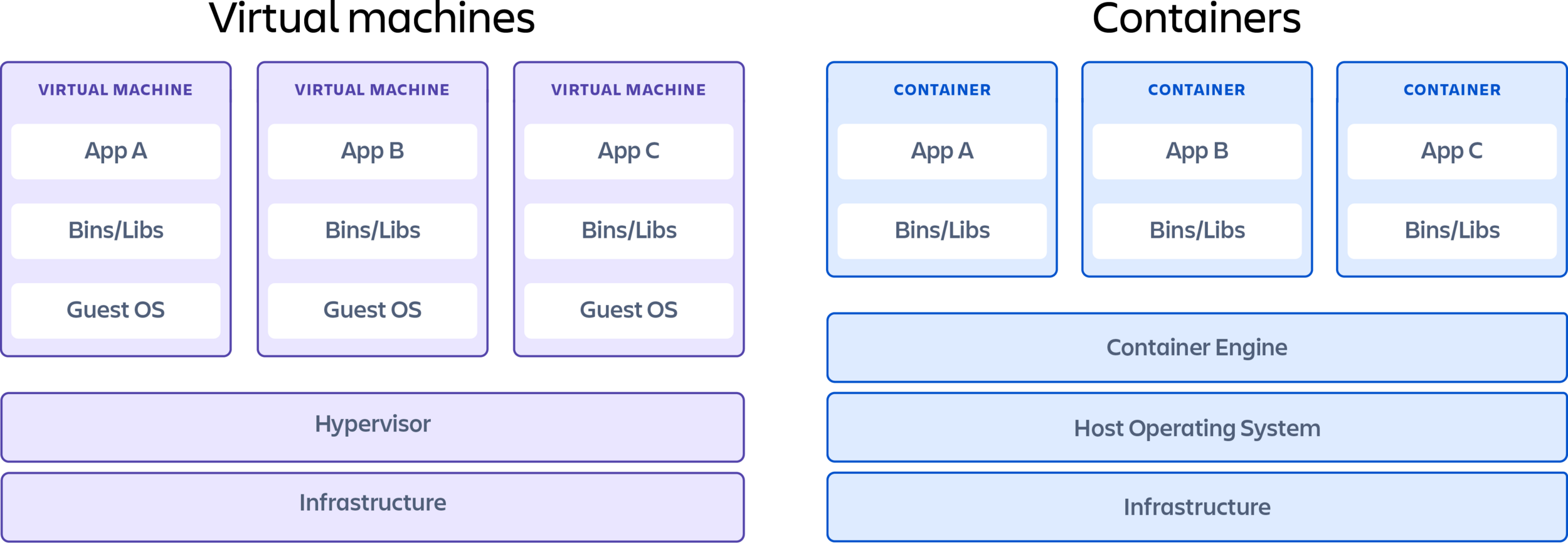
Containers vs VM:
Pros:
- Cost (less CPU, less memory)
- Faster runtime = faster runs
- Smaller sizes
- More portable*
Cons:
- Security (can be exploited)*
- Graphic User Interface is secondary*



Docker vs Apptainer
Docker:
- Targeting Laptops: better OS support (yes, you, mac/win peeps)
- Hosts public hub to share built containers
- Works with layer images to build containers
- Docker images can be used as bases for Apptainer recipes
Apptainer:
- Built for HPCs (Unix only), maintained by the Linux Foundation
- Easier "recipe" syntax
- Containers are a full root system folder that can be built as sandboxes
- Supports Docker images as bases
- It is safer than docker





Docker vs Apptainer


Bootstrap: docker
From: python:3.8.13-slim-buster
%files
.. /opt
%environment
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
%post
# Set install variables, create tmp folder
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
# Prepare repos and install dependencies
pip3 install /opt/.[all]
%runscript
nigsp
%labels
org.label-schema.name="NiGSP"
org.label-schema.description="NiGSP: python library for Graph Signal Processing on Neuroimaging data"
org.label-schema.url="https://github.com/miplabch/nigsp"
org.label-schema.vcs-url="https://github.com/miplabch/nigsp"
org.label-schema.schema-version="1.0"FROM python:3.8.13-slim-buster AS nigspdock
WORKDIR /app
# Prepare environment
COPY .. .
RUN pip3 install .[all]
ENV LANG="en_US.UTF-8" \
LC_ALL="en_US.UTF-8"
CMD nigsp
ARG BUILD_DATE
ARG VCS_REF
ARG VERSION
LABEL org.label-schema.build-date=$BUILD_DATE \
org.label-schema.name="NiGSP" \
org.label-schema.description="NiGSP: python library for Graph Signal Processing on Neuroimaging data" \
org.label-schema.url="https://github.com/miplabch/nigsp" \
org.label-schema.vcs-ref=$VCS_REF \
org.label-schema.vcs-url="https://github.com/miplabch/nigsp" \
org.label-schema.version=$VERSION \
org.label-schema.schema-version="1.0"
Docker
Apptainer
Example container: NiGSP


Bootstrap: docker
From: python:3.8.13-slim-buster
%environment
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
%post
# Set install variables, create tmp folder
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
# Prepare repos and install dependencies
pip3 install nigsp[all]
%runscript
nigsp
FROM python:3.8.13-slim-buster
# Prepare environment
ENV DEBIAN_FRONTEND=noninteractive \
TZ=Europe/Brussels
RUN pip3 install nigsp[all]
CMD nigsp
Docker
Apptainer
# Create a sandbox image from a python image on the Docker Hub
apptainer build --fakeroot --sandbox nigsp.img docker://python:3.8.13-slim-buster
# Start an interactive session to modify the sandbox (short and long flags)
apptainer shell -f -e -w --no-home nigsp.img
apptainer shell --fakeroot --cleanenvironment --writable --no-home nigsp.img
# Install nigsp in the container
pip3 install nigsp[all]
pip3 list
# Exit the container
exit
# Create an unmodifiable image from the previous sandbox
apptainer build -f nigsp.sif nigsp.img
# Execute a command in the new image, in this case call the help of nigsp
apptainer exec -e --no-home nigsp.sif nigsp --helpApptainer example: complete data analysis.


Bootstrap: docker
From: ubuntu:{{ UBUNTU_VER}}
%arguments
UBUNTU_VER=22.04
R_VER=4.3.2-1.2204.0
AFNI_VER=24.0.00
ANTS_VER=2.5.0
C3D_VER=1.0.0
FSL_VER=6.0.7.6
P2C_VER=0.18.3
PK_VER=0.2.1
EUSKPREPROC_VER=0.7.1
%environment
# export templateloc=/usr/share/afni/atlases
export AFNIPATH="/opt/afni-AFNI_{{ AFNI_VER }}"
export AFNI_PLUGINPATH="$AFNIPATH"
export templateloc=/usr/share/afni/atlases
export AFNI_AUTOGZIP=YES
export AFNI_COMPRESSOR=GZIP
export ANTSPATH="/opt/ants-{{ ANTS_VER }}/bin"
export ANTSSCRIPTS="/opt/ants-{{ ANTS_VER }}/Scripts"
export C3DPATH="/opt/convert3d-{{ C3D_VER }}"
export FSLDIR="/opt/fsl-{{ FSL_VER }}"
source ${FSLDIR}/etc/fslconf/fsl.sh
export FSLOUTPUTTYPE="NIFTI_GZ"
export FSLMULTIFILEQUIT="TRUE"
export FSLTCLSH="$FSLDIR/bin/fsltclsh"
export FSLWISH="$FSLDIR/bin/fslwish"
export FSLLOCKDIR=""
export FSLMACHINELIST=""
export FSLREMOTECALL=""
export FSLGECUDAQ="cuda.q"
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
export R_LIBS="/usr/lib/R"
export LD_LIBRARY_PATH="/opt/ants-{{ ANTS_VER }}/lib:$LD_LIBRARY_PATH"
export PREPROCPATH="/opt/preprocessing"
export PATH="$AFNIPATH:$ANTSPATH:$ANTSSCRIPTS:$C3DPATH/bin:$FSLDIR/bin:$PREPROCPATH:$PREPROCPATH/00.pipelines:$PATH"
%post
# Set install variables, create tmp folder
export TMPDIR="/tmp/general_preproc_build_$( date -u +"%F_%H-%M-%S" )"
[[ -d ${TMPDIR} ]] && rm -rf ${TMPDIR}
mkdir -p ${TMPDIR}
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
apt update -qq
apt install -y -q --no-install-recommends ca-certificates dirmngr gnupg
# Prepare repos and install dependencies
apt-key adv --keyserver keyserver.ubuntu.com --recv-keys C9A7585B49D51698710F3A115E25F516B04C661B
apt-key adv --keyserver keyserver.ubuntu.com --recv-keys 6E12762B81063D17BDDD3142F142A4D99F16EB04
echo "deb https://ppa.launchpadcontent.net/marutter/rrutter4.0/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb-src https://ppa.launchpadcontent.net/marutter/rrutter4.0/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb https://ppa.launchpadcontent.net/c2d4u.team/c2d4u4.0+/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb-src https://ppa.launchpadcontent.net/c2d4u.team/c2d4u4.0+/ubuntu focal main" | tee -a /etc/apt/sources.list
apt update -qq
apt install -y -q --no-install-recommends \
bc \
build-essential \
bzip2 \
cmake \
curl \
dc \
file \
freeglut3-dev \
g++ \
gcc \
git \
less \
libcurl4-openssl-dev \
libeigen3-dev \
libexpat1-dev \
libf2c2-dev \
libfftw3-3 \
libfftw3-dev \
libgdal-dev \
libgfortran4 \
libgfortran-8-dev \
libglew-dev \
libgl1-mesa-dev \
libgl1-mesa-dri \
libgl1-mesa-glx \
libglib2.0-dev \
libglu1-mesa-dev \
libglw1-mesa \
libgomp1 \
libgsl-dev \
libgts-dev \
libjpeg8-dev \
liblapack3 \
libopenblas-dev \
libmotif-dev \
libnetpbm10-dev \
libnode-dev \
libpng16-16 \
libpng-dev \
libquadmath0 \
libtiff5 \
libtiff5-dev \
libudunits2-dev \
libxext-dev \
libxi-dev \
libxm4 \
libxmhtml-dev \
libxml2-dev \
libxmu-dev \
libxmu-headers \
libxpm-dev \
libxt-dev \
m4 \
make \
mesa-common-dev \
nano \
r-base-dev \
rsync \
tcsh \
python3-distutils \
python3-pip \
python3-rpy2 \
python-is-python3 \
qhull-bin \
xvfb \
zlib1g-dev
# Install AFNI
mkdir -p ${TMPDIR}/afni
cd ${TMPDIR}/afni || exit 1
ln -s /usr/lib/x86_64-linux-gnu/libgsl.so.23 /usr/lib/x86_64-linux-gnu/libgsl.so.19
ln -s /usr/lib/x86_64-linux-gnu/libXp.so.6 /usr/lib/x86_64-linux-gnu/libXp.so
git clone https://github.com/afni/afni.git source
cd source || exit 1
git fetch --tags
git -c advice.detachedHead=false checkout AFNI_22.3.07
cd src || exit 1
cp other_builds/Makefile.linux_ubuntu_16_64_glw_local_shared Makefile
make itall
mv linux_ubuntu_16_64_glw_local_shared /opt/afni-AFNI_22.3.07
export PATH="/opt/afni-AFNI_22.3.07:$PATH"
export R_LIBS="/usr/lib/R"
rPkgsInstall -pkgs ALL
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/afni
# Install ANTs
mkdir -p ${TMPDIR}/ants/build
git clone https://github.com/ANTsX/ANTs.git ${TMPDIR}/ants/source
cd ${TMPDIR}/ants/source || exit 1
git fetch --tags
git -c advice.detachedHead=false checkout v2.4.2
cd ${TMPDIR}/ants/build || exit 1
cmake -DCMAKE_INSTALL_PREFIX=/opt/ants-2.4.2 -DBUILD_SHARED_LIBS=ON -DBUILD_TESTING=OFF ${TMPDIR}/ants/source
make -j 10
mkdir -p /opt/ants-2.4.2
cd ANTS-build || exit 1
make install
mv ../../source/Scripts/ /opt/ants-2.4.2
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/ants
# Install C3D
echo "Downloading Convert3D ..."
mkdir -p /opt/convert3d-1.0.0
curl -fsSL https://sourceforge.net/projects/c3d/files/c3d/1.0.0/c3d-1.0.0-Linux-x86_64.tar.gz/download \
| tar -xz -C /opt/convert3d-1.0.0 --strip-components 1
# Install FSL
mkdir -p ${TMPDIR}/fsl
cd ${TMPDIR}/fsl || exit 1
curl -fL https://fsl.fmrib.ox.ac.uk/fsldownloads/fslinstaller.py --output ./fslinstaller.py
chmod +x fslinstaller.py
python3 fslinstaller.py -d /opt/fsl-6.0.6.2 -V 6.0.6.2
# echo "Installing FSL conda environment ..."
# bash /opt/fsl-6.0.6.2/etc/fslconf/fslpython_install.sh -f /opt/fsl-6.0.6.2
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/fsl
# Clone EuskalIBUR preprocessing.
git clone https://github.com/smoia/EuskalIBUR_preproc.git /opt/preprocessing
apt install -y -q csvtool
# Install PYTHON things.
pip3 install pip==22.3.1 setuptools==65.5.1 wheel==0.38.4
# Install wxPython in a particular way.
pip3 install --no-cache -f https://extras.wxpython.org/wxPython4/extras/linux/gtk3/ubuntu-20.04 wxpython==4.2.0
# Install datalad, fsleyes, nilearn, peakdet, phys2cvr.
pip3 install \
annexremote==1.6.0 \
boto==2.49.0 \
certifi==2022.12.7 \
cffi==1.15.1 \
chardet==4.0.0 \
charset-normalizer==2.1.1 \
contourpy==1.0.6 \
cryptography==38.0.4 \
cycler==0.11.0 \
datalad==0.17.10 \
dill==0.3.6 \
distro==1.8.0 \
fasteners==0.18 \
fonttools==4.38.0 \
fsleyes==1.5.0 \
fsleyes-props==1.8.2 \
fsleyes-widgets==0.12.3 \
fslpy==3.10.0 \
h5py==3.7.0 \
humanize==4.4.0 \
idna==3.4 \
importlib-metadata==5.1.0 \
iso8601==1.1.0 \
jaraco.classes==3.2.3 \
jeepney==0.8.0 \
Jinja2==3.1.2 \
joblib==1.2.0 \
keyring==23.11.0 \
keyrings.alt==4.2.0 \
kiwisolver==1.4.4 \
lxml==4.9.2 \
MarkupSafe==2.1.1 \
matplotlib==3.6.2 \
more-itertools==9.0.0 \
msgpack==1.0.4 \
nibabel==4.0.2 \
nilearn==0.9.2 \
numpy==1.23.5 \
packaging==22.0 \
pandas==1.5.2 \
patool==1.12 \
peakdet==0.2.0rc1 \
phys2cvr==0.16.0 \
Pillow==9.3.0 \
platformdirs==2.6.0 \
pycparser==2.21 \
PyOpenGL==3.1.6 \
pyparsing==2.4.7 \
python-dateutil==2.8.2 \
python-gitlab==3.12.0 \
pytz==2022.6 \
requests==2.28.1 \
requests-toolbelt==0.10.1 \
scikit-learn==1.2.0 \
scipy==1.9.3 \
SecretStorage==3.3.3 \
simplejson==3.18.0 \
six==1.16.0 \
threadpoolctl==3.1.0 \
tqdm==4.64.1 \
urllib3==1.26.13 \
Whoosh==2.7.4 \
zipp==3.11.0
# Final removal of lists and cleanup
cd /tmp || exit 1
rm -rf ${TMPDIR}
rm -rf /var/lib/apt/lists/*Easy containers
Containers in action
apptainer build --sandbox container.img docker://afni/afni_dev_base:AFNI_22.2.12
apptainer shell -f -e -w --no-home container.img
apptainer build container.sif recipe.def
apptainer exec -f -e --no-home -B /some/place:/tmp
-B /some/place/elsewhere:/scripts \
-B /another/place/:/data \
container.sif /scripts/run_batch_analysis.sh sub-001 ses-01apptainer exec docker://ghcr.io/apptainer/lolcow cowsay "Hello $USER!"Try it now yourselves!



Practical #1
- Get the phys2cvr test data
- Write a recipe to install phys2cvr in an Ubuntu 22.04 container
- Build the container
- Run phys2cvr in that container



Practical #1



https://files.de-1.osf.io/v1/resources/mcr8g/providers/osfstorage/?zip=Practical #1



Bootstrap: docker
From: ubuntu:22.04
%environment
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
%post
# Set install variables, create tmp folder
export TMPDIR="/tmp/general_preproc_build_$( date -u +"%F_%H-%M-%S" )"
[[ -d ${TMPDIR} ]] && rm -rf ${TMPDIR}
mkdir -p ${TMPDIR}
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
apt update -qq
apt install -y -q --no-install-recommends ca-certificates dirmngr gnupg lsb-release wget
apt install -y -q --no-install-recommends python3-distutils python3-pip python-is-python3
# Install PYTHON things.
pip3 install pip==25.0 setuptools==70.3.0 wheel==0.37.1
# Install datalad, fsleyes, nilearn, peakdet, phys2cvr.
pip3 install phys2cvr==0.18.6
# Final removal of lists and cleanup
cd /tmp || exit 1
rm -rf ${TMPDIR}
rm -rf /var/lib/apt/lists/*
Practical #1



apptainer build -f p2c.sif recipe_p2c.def
apptainer shell -f -e --no-home -B ~/mydata:/data p2c.sif
cd /data
phys2cvr -i func.nii.gz -o results -m mask.nii.gz -r roi.nii.gz -co2 co2.phys -dmat motpar.par motderiv.par
apptainer exec -f -e --no-home -B /some/place:/tmp
-B /some/place/elsewhere:/scripts \
-B /another/place/:/data \
p2c.sif phys2cvr -i /data/func.nii.gz -o /data/results \
-m /data/mask.nii.gz -r /data/roi.nii.gz -co2 /data/co2.phys \
-dmat /data/motpar.par /data/motderiv.par3. Version Control Systems



Does any of these situations look familiar?
I can't work on that project now because my colleague/friend/dog is working on [a different part than what I'd modify of] it at the moment...



Version Control Systems (VCS)
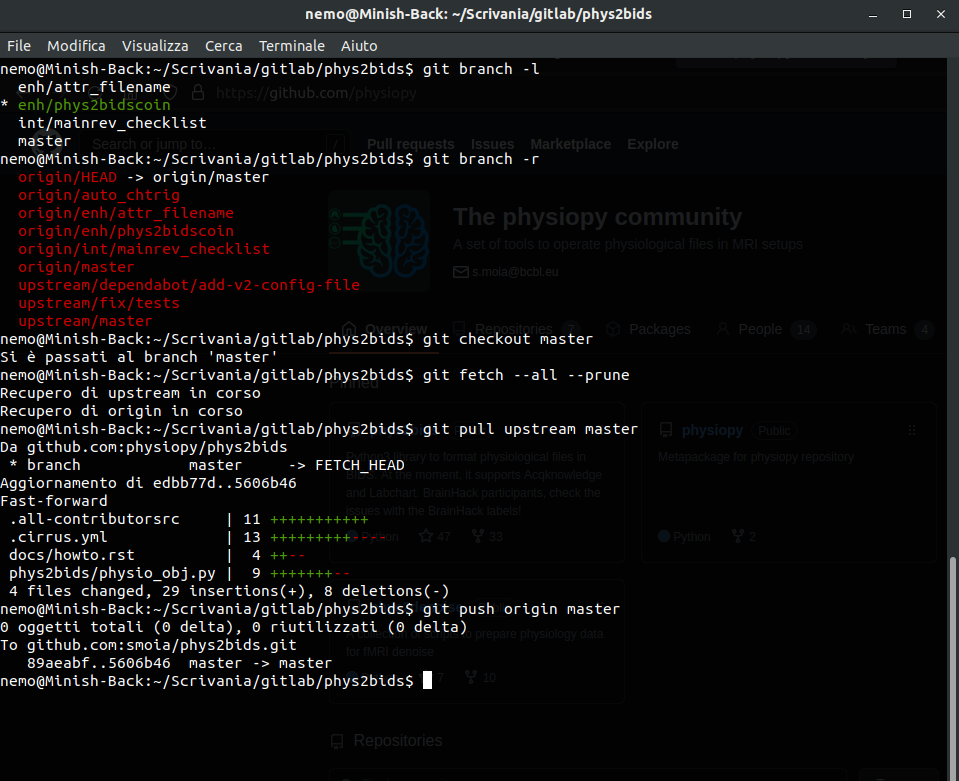



Version Control Systems (VCS)
Version Control
Version control systems are a way to manage and track changes to files.







Content
Aggregation/delivery



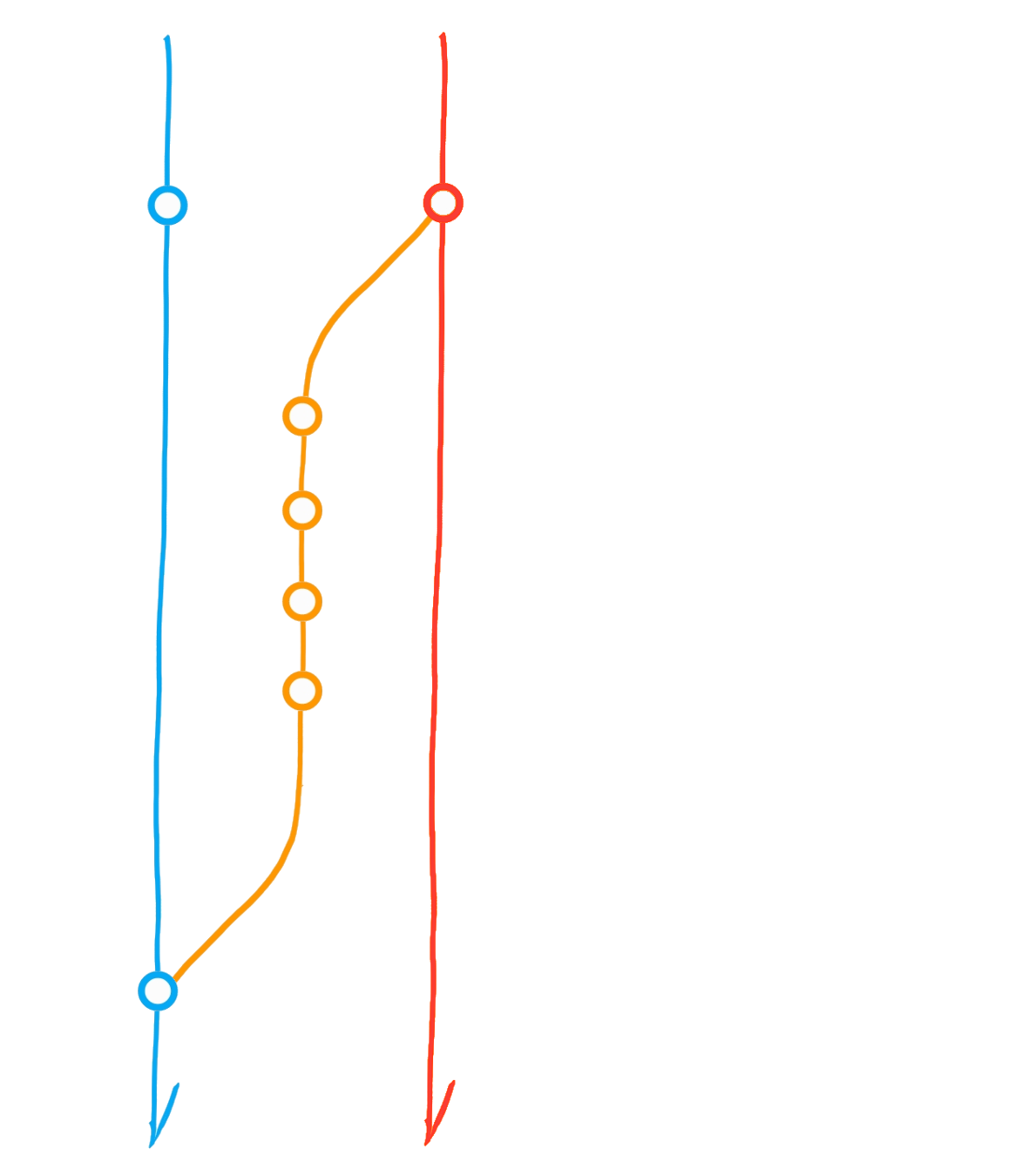
A classic git(Hub) flow

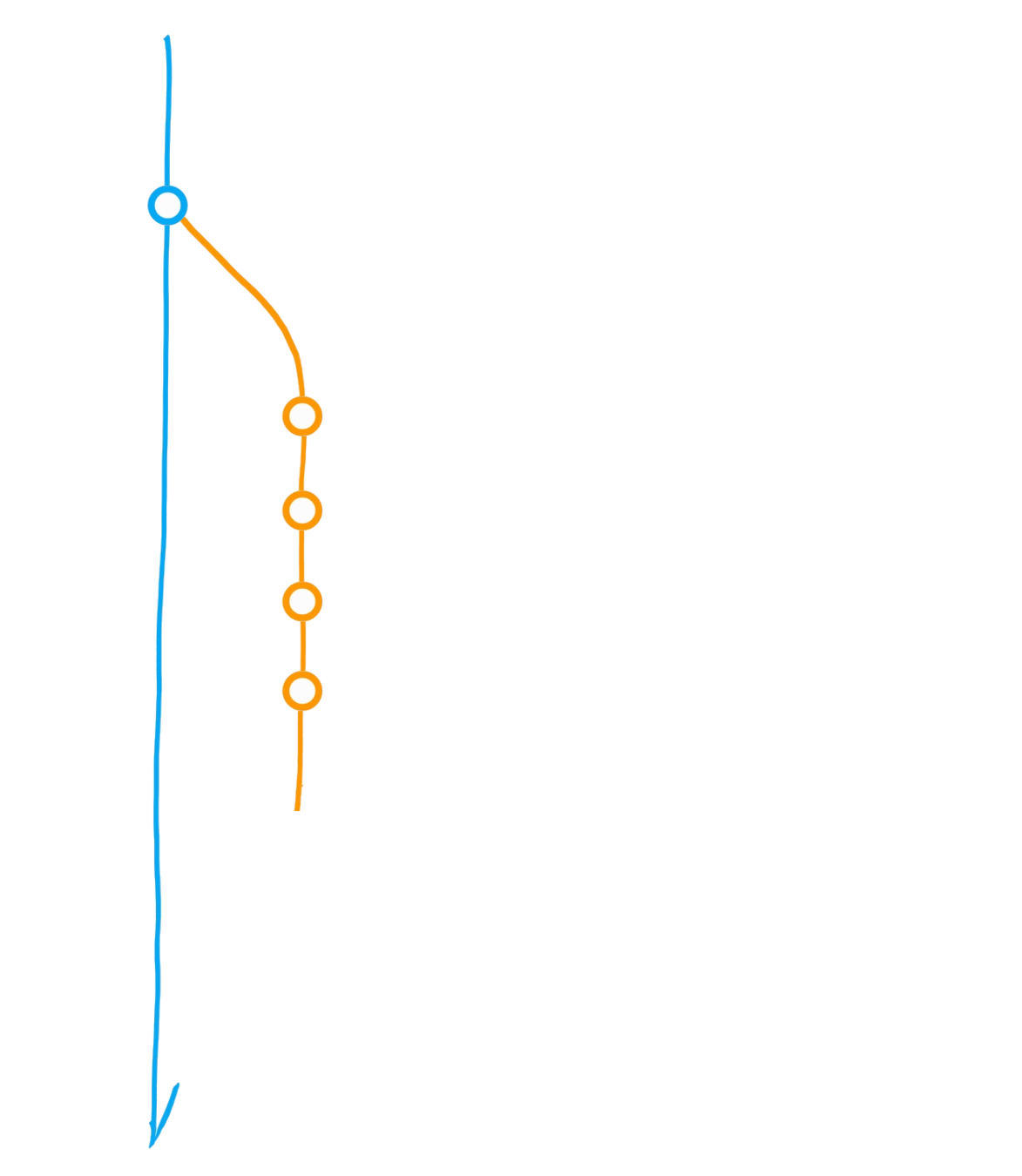
Create branch "dev"
Commit
Merge dev into main
Diverging main: conflict?
Merge main into dev
Initialise repository
"Main" branch
Main
Dev
Bug



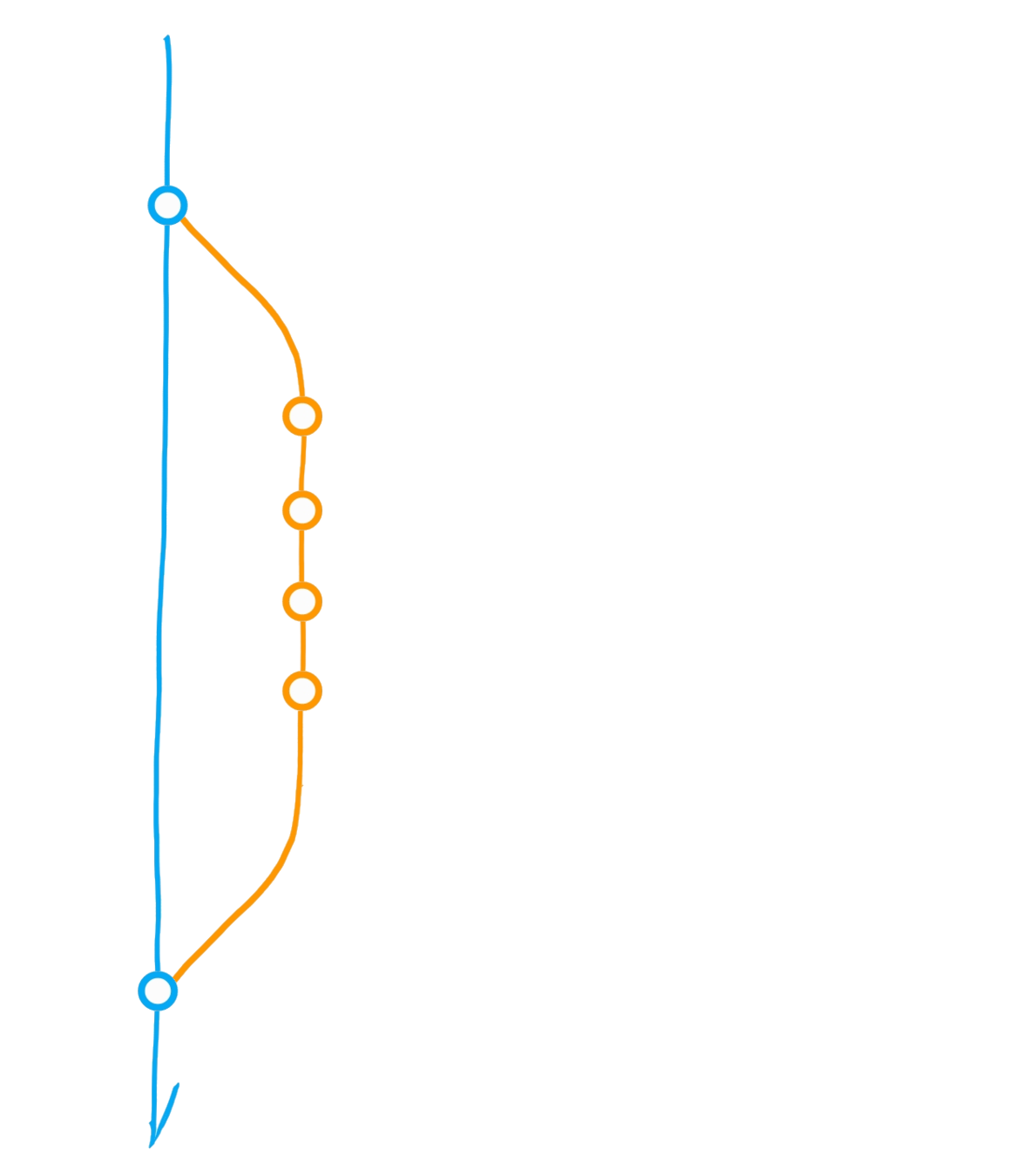
A classic git(Hub) flow
Create branch "dev"
Commit
Merge dev into main
Diverging main: conflict?
Merge main into dev
Initialise repository
"Main" branch

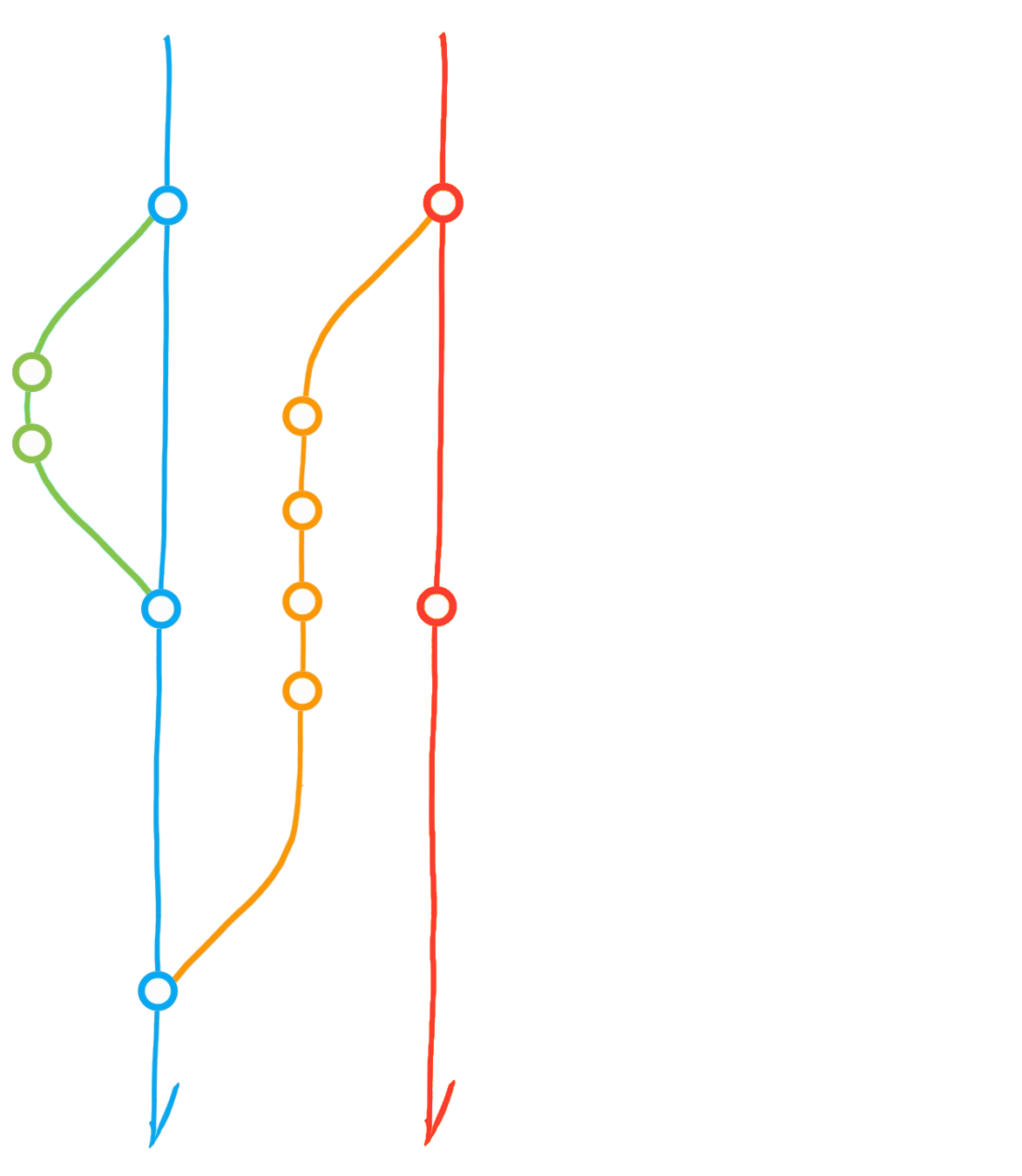

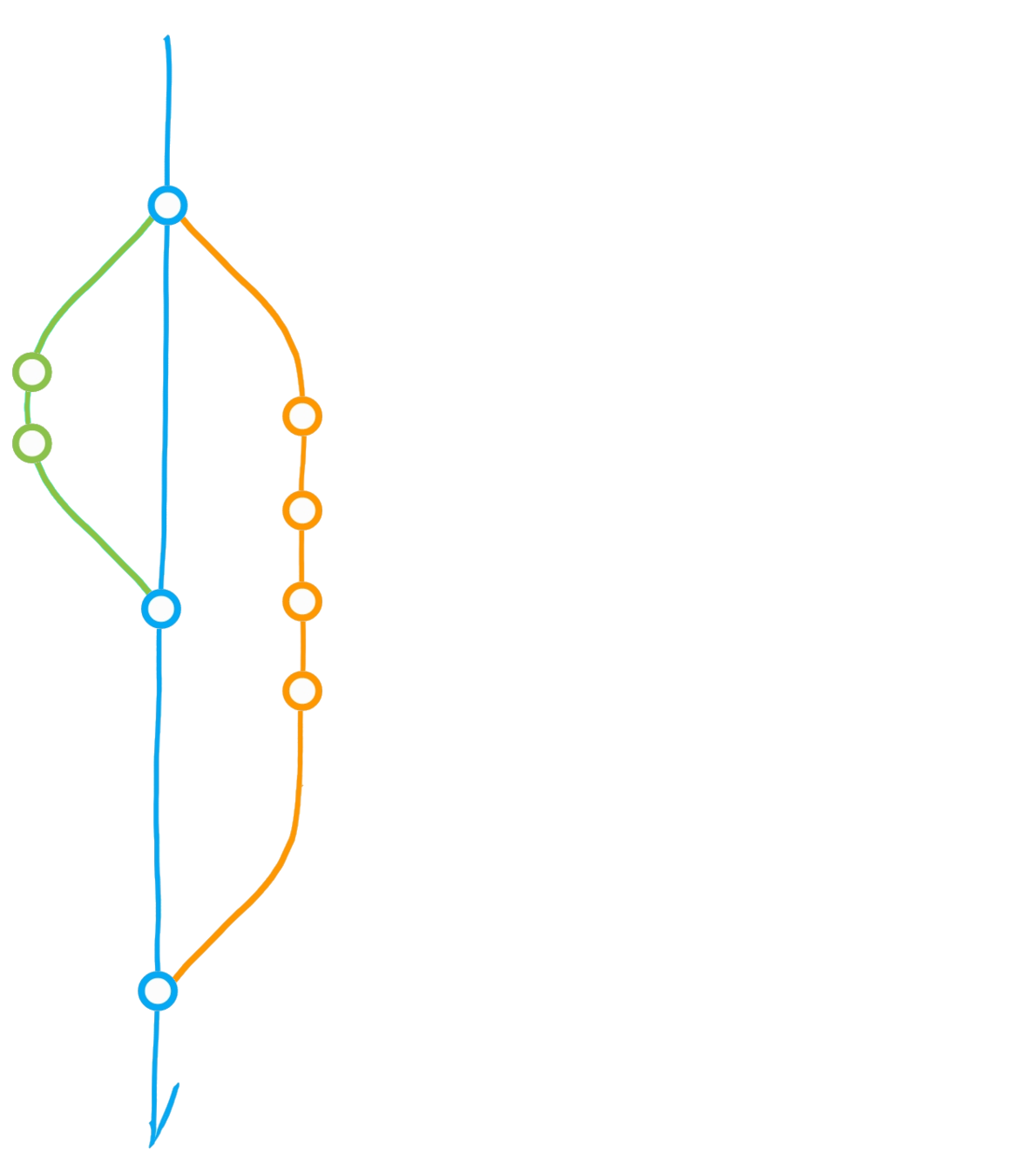
Fork ("upstream" vs "origin")
Pull from upstream
Merge origin/main into dev
Clone (local repository)
Pull Request
Pull from *
Push to *


Main
Dev
Upstream
Main
Origin
Dev
Main
(local)




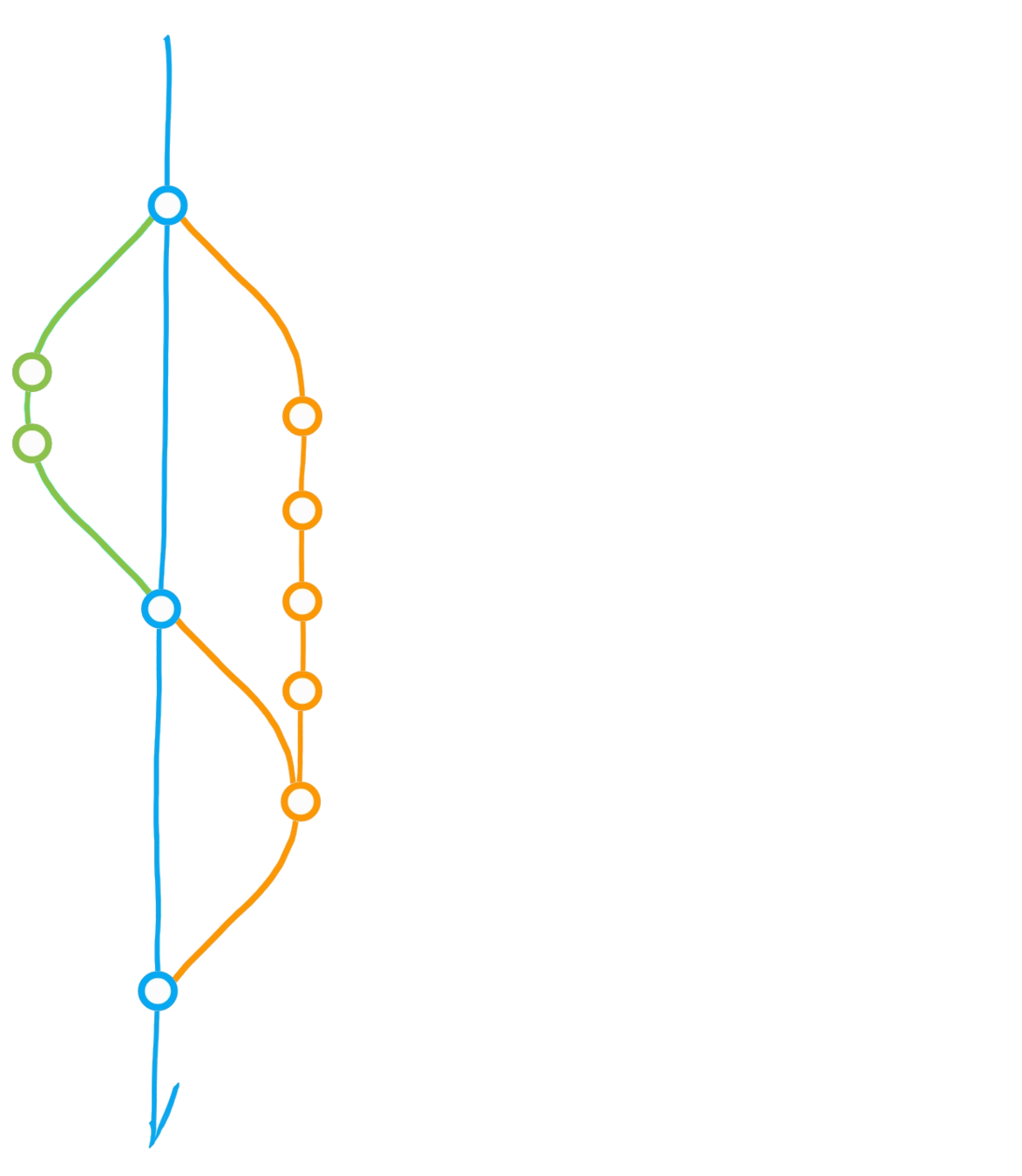
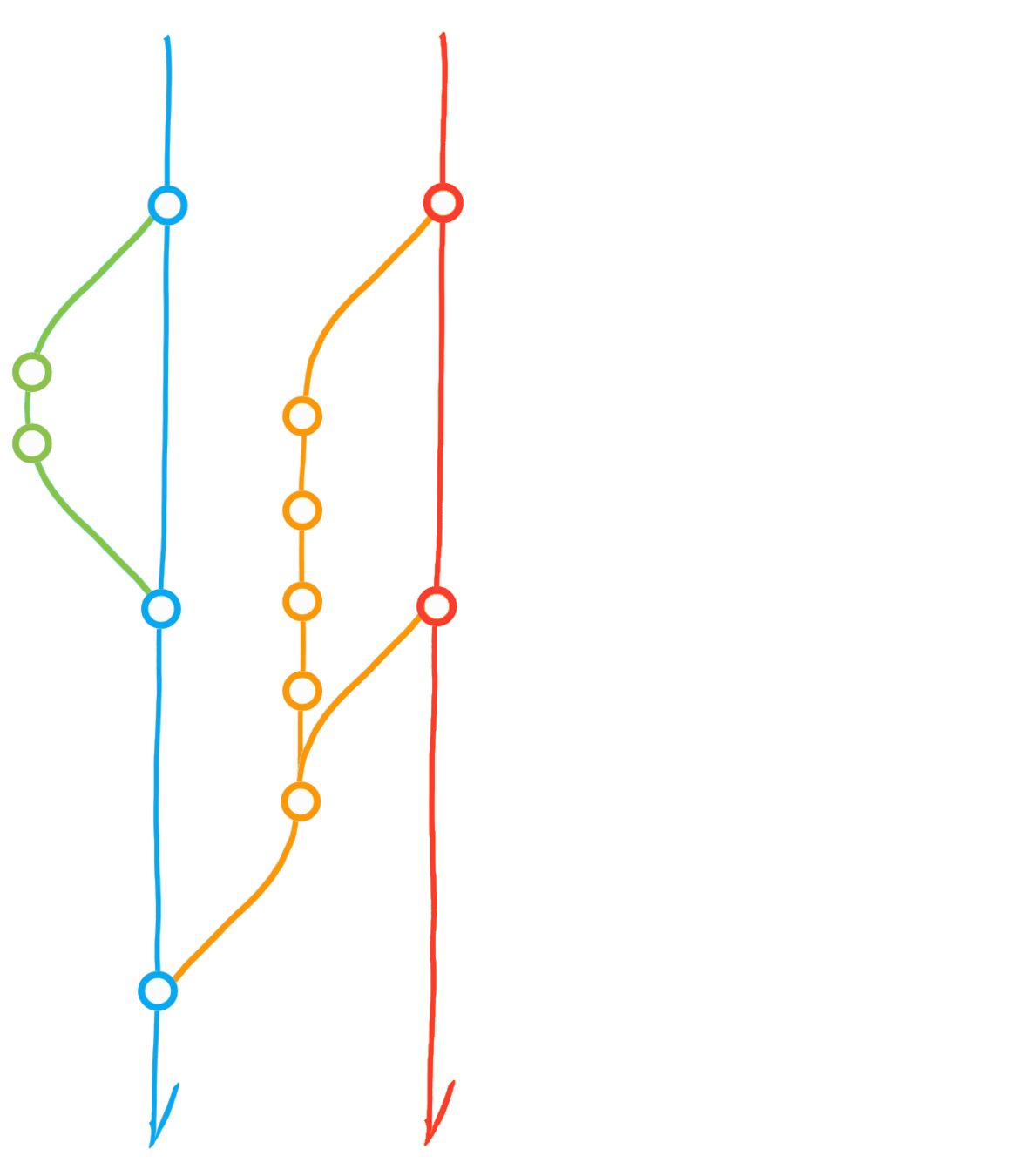
File history & parallel development
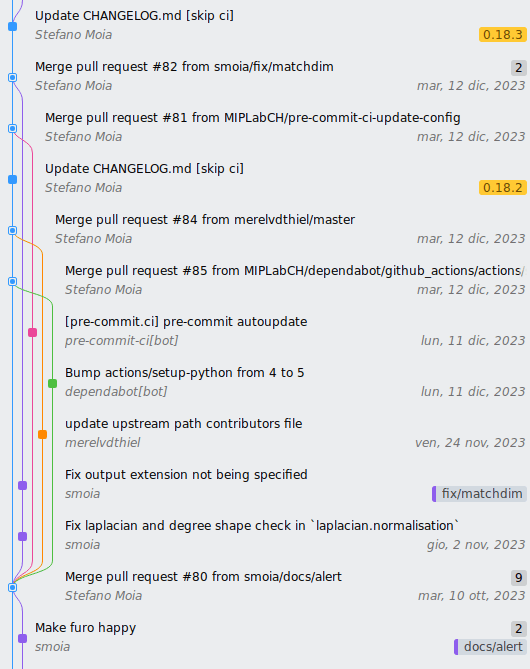
Attribution
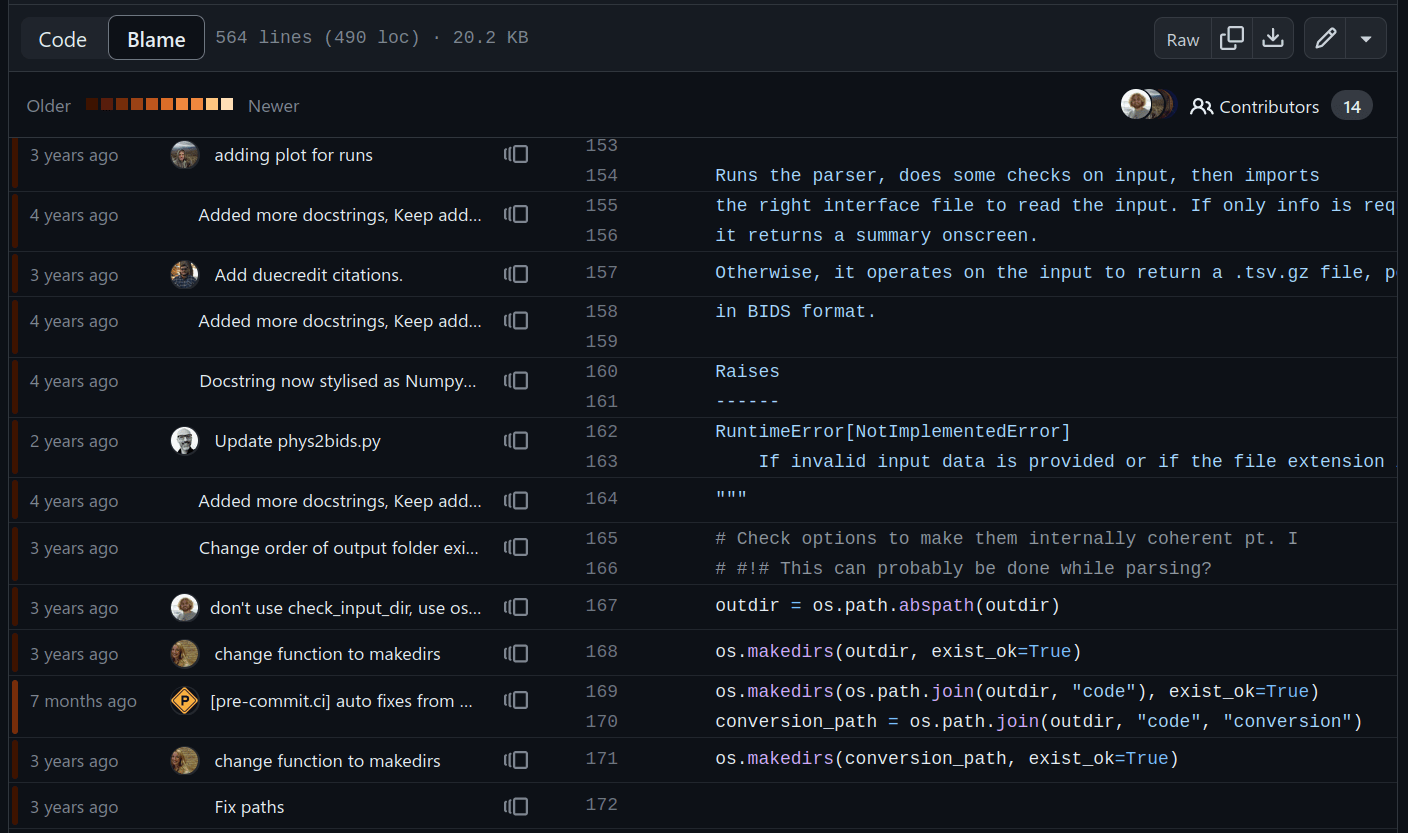






Automation pt. 1: git hooks
pip install pre-commit # Install via pip, or
# Comes installed with development extras
pip install -e /path/to/phys2cvr[dev]
cd /path/to/phys2cvr
pre-commit init
pre-commit run(Local and remote) simple automations, e.g:
- Code style
- File checks (empty lines, indent, executables)
- Language and typos (!!!)



Pull requests and Reviews
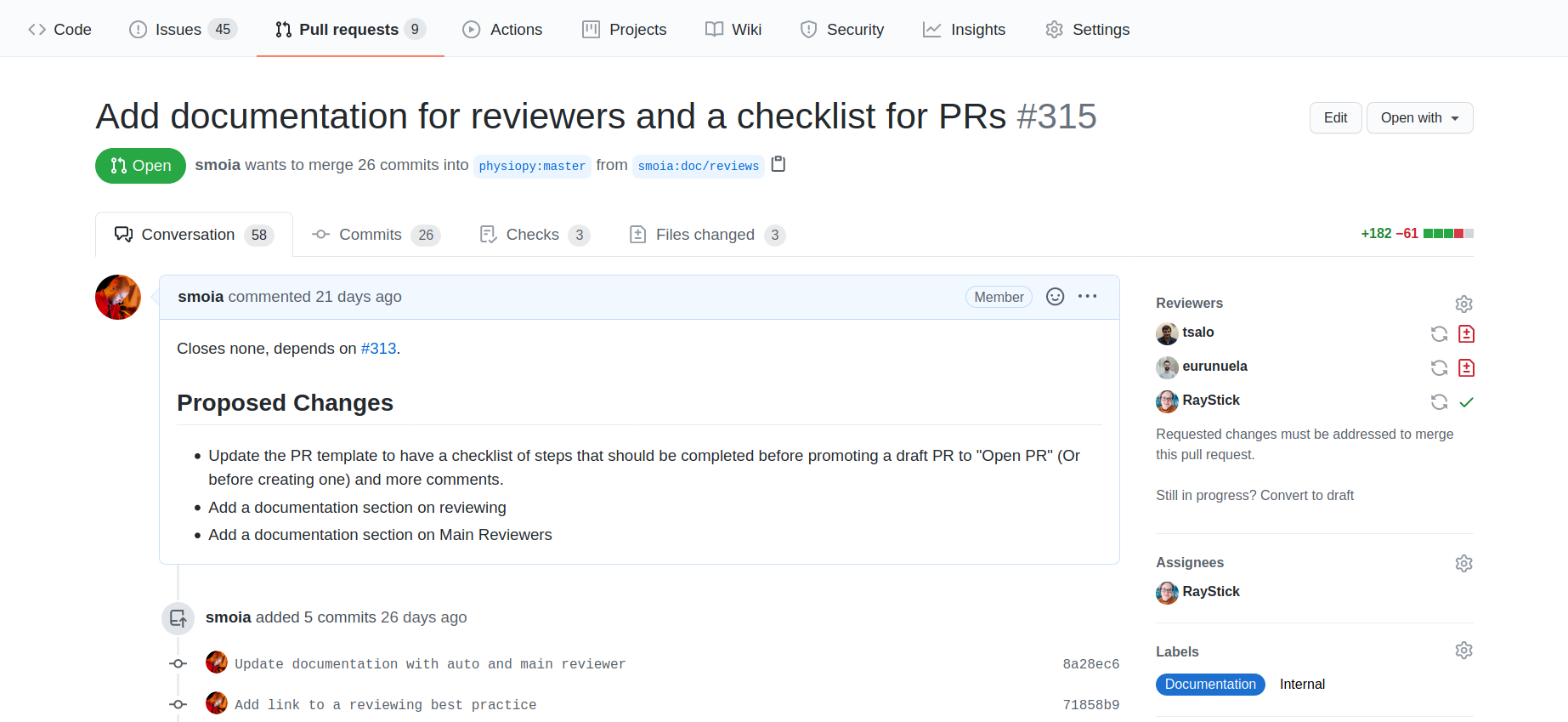






Pull requests and Reviews
Some suggestions for...
- Keep you contribution small and focused
- Make your contribution as clear as possible
- Use a review as a learning experience
- Be patient: reviewers might ask you some more work than you expected, but it's always to improve your work.
- Be kind and patient
- Don't limit your review to the apparent changes - depending on the importance of the review, take the time to look at how the whole project might change.
- Keep your review to what's necessary for the contribution - if it would be nice to ..., open an issue (or think about making the contribution yourself).
... Authors
... Reviewers



Take home #2
Working with git(Hub) allows you to:
-
work in parallel on new features without disrupting the "main" version of your project
-
track changes in time.
Bonus: it can force a team to double check projects!



Practical #2.1
- Configure git
- Create a folder and put a LICENCE and the recipe you created there
- Initialise that folder as a git repository
- Create a new public repository on GitHub (same name as your folder!)
- Push your local folder to that GitHub repository
- Take a moment to appreciate your first GitHub interaction!



Practical #2.2
- Fork your collaborator's repository (upstream) to create your own (origin)
- Clone origin
- Create a new branch and add the installation of phys2bids in the container definition
- Push your local changes to origin
- Open a PR to upstream
- Look at the PR you received, comment it, and if it's ok, approve it. If not, take your time to (respectfully) discuss what you would do different.
Remember: PRs are public conversations, following etiquette!!!



Practical #2.3
- Once you merged your PR, take a moment to celebrate your first merged PR!
- Then show me the whole process.



4. Licenses



Disclaimer:
I am not a legal expert.
If you ever have any doubts, contact the Technology Transfer Office.



License your work
A work that is not licensed is not public (paradox!)
There are n+1 (open source) licences to pick up from.
www.choosealicense.org
The licence should be the first commit you make in a project.
Personal picks for science:
Apache 2.0 and CC-BY-ND-4.0
(consider L-GPLv3.0, and CC-BY-4.0 too)



Understand licensing and ownership
- Check the licence of code, data, and libraries you are "borrowing".
- Pay attention to single vs double licensing (e.g. academic vs commercial).
- Check licence compatibility.
- Remember that institutions might have rights to what their employees do:
- However, they can also help you with licensing and license enforcement.
The data resulting from the research, as well as the publication rights are owned by the Host Institute [UM], unless otherwise stated below in the section “Additional arrangements”.



Licence compatibility
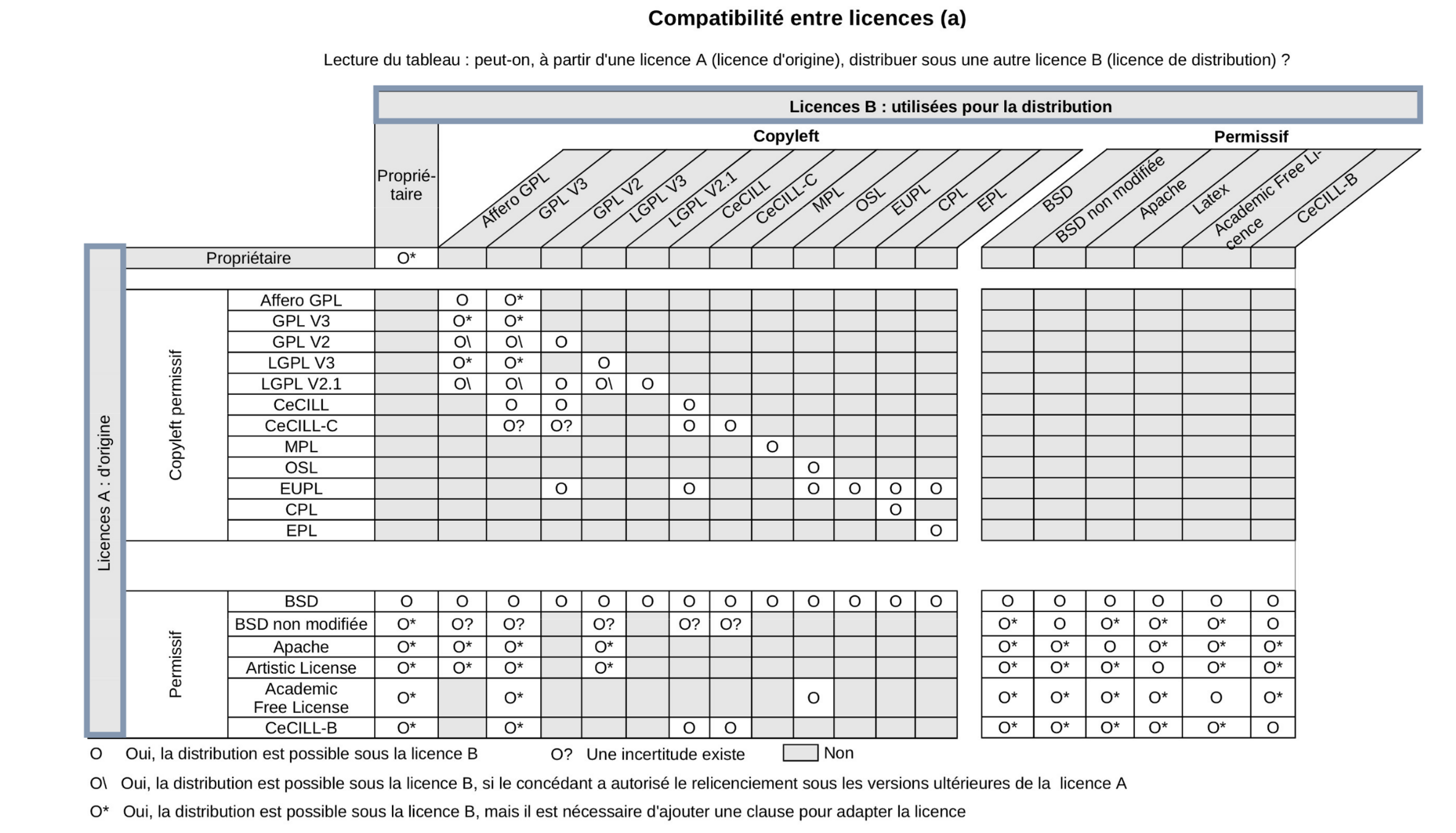
© Sebastien Adams, I WANT TO DISTRIBUTE MY SOFTWARE DEVELOPMENTS. HOW TO DEFINE AN OPEN LICENSING STRATEGY?
© Benjamin Jean (2011), Option libre. Du bon usage des licences libres.
License your work in the right way
- Put a copy of the licence or a link to it as close as possible to "borrowed" material, if not in it.
- If any license requires its adoption for derivatives (e.g. GPL), you must licence your work with the same licence.
- You can ask the original authors to change their licence (e.g. GPL to L-GPL) or give you special permissions.
- Remember to add licences disclaimers in all of your files.
[...]
if __name__ == "__main__":
_main(sys.argv[1:])
"""
Copyright 2022, Stefano Moia & EPFL.
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
"""



License your work in the right way

[...]



License your work in the right way
MATLAB users:
- If you include external functions/scripts/libraries, your work is considered a derivative. Report licence, authors, and origin of the code inside them and respect their licence.
- Alternatively, don't include anything but state requirements / create install scripts.
- If you are releasing a build, the build is considered a derivative.
Python users:
- If you copy-paste code, your work is a derivative.
- Imports are trickier:
- Technically, GPL or © licences triggers on import.
- Practically, it's a really grey area. Make those imports optional, and specify their licences as clearly as possible.



Take home #2
Licensing is as complicated as it is important.
Double check licenses of borrowed material, report them in your own work
for licence tracking.
Remember that institutions have rights on your work, but can should help you with licensing.
Any question [/opinions/objections/...]?

Practical intro to computationally reproducible science [UM]
By Stefano Moia
Practical intro to computationally reproducible science [UM]
CC-BY 4.0 Stefano Moia, 2025. Images are property of the original authors and should be shared following their respective licences. This presentation is otherwise licensed under CC BY 4.0. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/
- 199



