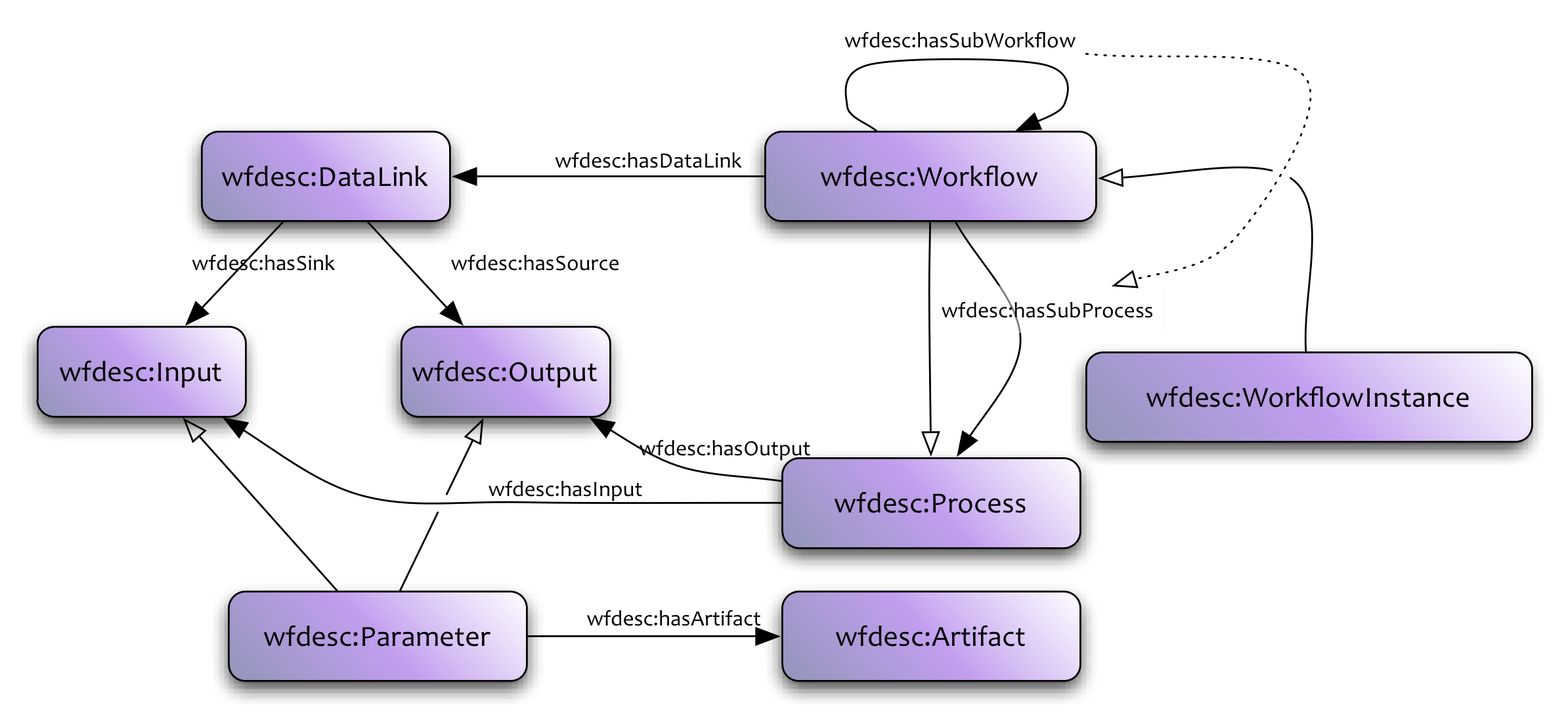
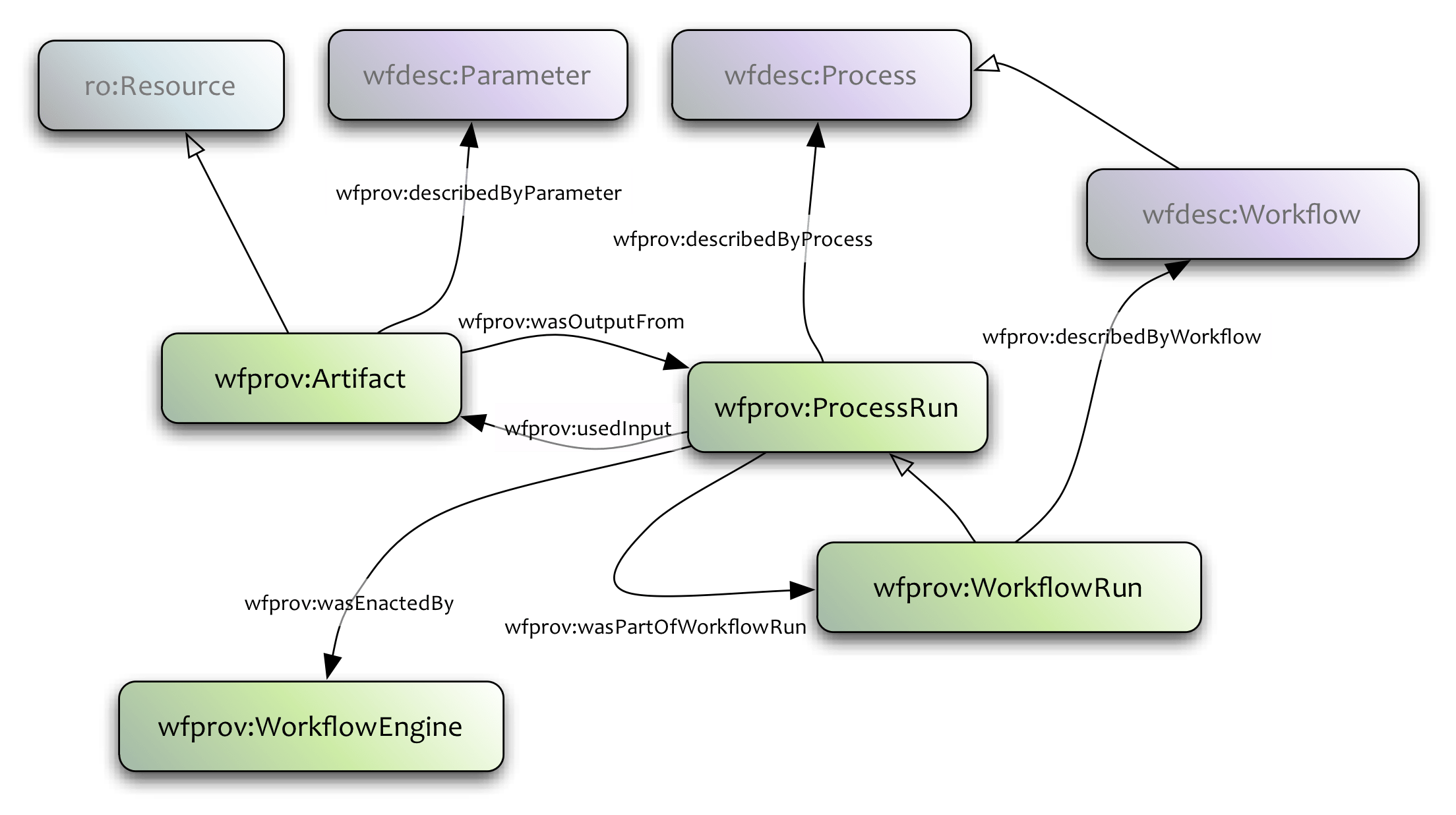
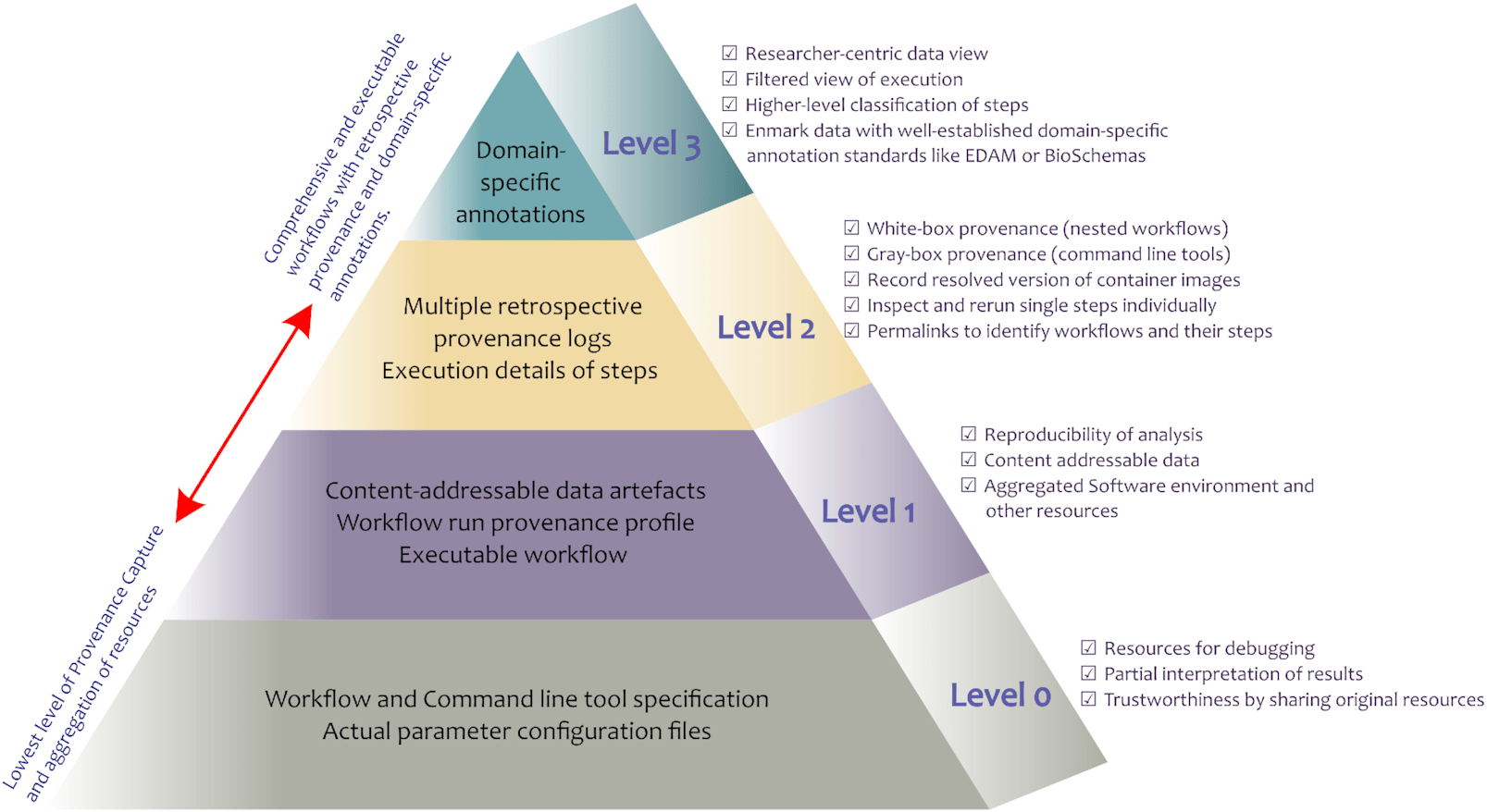
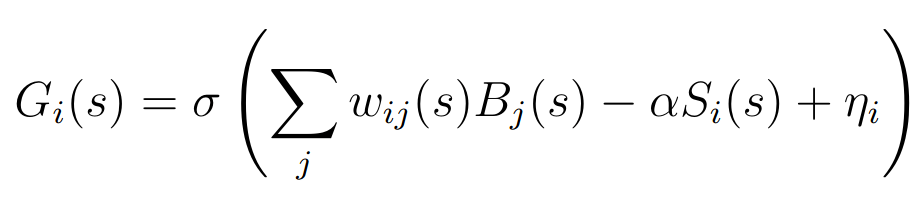<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Leadóg"@ga .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Tennis"@sv .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Tenis"@es .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "テニス"@ja .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Tenis"@cs .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "网球"@zh .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Теннис"@ru .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Tenis"@pl .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Tennis"@de .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Teniso"@eo .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Tennis"@en .
<http://dbpedia.org/resource/Tennis> <http://www.w3.org/2000/01/rdf-schema#label> "Tenis"@eu .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/ontology/abstract> "Tennis is a racket sport that is played either individually against a single opponent (singles) or between two teams of two players each (doubles). Each player uses a tennis racket that is strung with cord to strike a hollow rubber ball covered with felt over or around a net and into the opponent's court. The object of the game is to manoeuvre the ball in such a way that the opponent is not able to play a valid return. The player who is unable to return the ball validly will not gain a point, while the opposite player will. Tennis is an Olympic sport and is played at all levels of society and at all ages. The sport can be played by anyone who can hold a racket, including wheelchair users. The modern game of tennis originated in Birmingham, England, in the late 19th century as lawn tennis. It had close connections both to various field (lawn) games such as croquet and bowls as well as to the older racket sport today called real tennis. The rules of modern tennis have changed little since the 1890s. Two exceptions are that until 1961 the server had to keep one foot on the ground at all times, and the adoption of the tiebreak in the 1970s. A recent addition to professional tennis has been the adoption of electronic review technology coupled with a point-challenge system, which allows a player to contest the line call of a point, a system known as Hawk-Eye. Tennis is played by millions of recreational players and is a popular worldwide spectator sport. The four Grand Slam tournaments (also referred to as the majors) are especially popular: the Australian Open, played on hardcourts; the French Open, played on red clay courts; Wimbledon, played on grass courts; and the US Open, also played on hardcourts."@en .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/ontology/abstract> "Ténis (português europeu) ou tênis (português brasileiro) é um esporte de origem inglesa, disputado em quadras geralmente abertas e de superfícies sintéticas, cimento, saibro ou relva. Participam no jogo dois oponentes ou duas duplas de oponentes, podendo ser mistas (homens e mulheres) ou não. A quadra é dividida em duas meia-quadras por uma rede, e o objetivo do jogo é rebater uma pequena bola para além da rede (para a meia-quadra adversária) com ajuda de uma raquete. Para marcar um ponto é preciso que a bola toque no solo em qualquer parte dentro da quadra adversária incluindo as linhas que demarcam o campo do oponente, fazendo com que o adversário não consiga devolver a bola antes do segundo toque, ou que a devolva para fora dos limites da outra meia-quadra. O desporto assim possui aspectos de ataque (rebater bem a bola, dificultando a devolução do adversário) e defesa (bom posicionamento em quadra, antecipação do lance adversário etc). O tênis possui um intricado sistema de pontuação, que subdivide o jogo em games/jogospt e sets/partidas pt. Grosso modo, um game é um conjunto de pontos (15-30-40-game) e um set é um conjunto de games (1-2-3-4-5-set). Cada game tem um jogador responsável por recolocar a bola em jogo: fazer o serviço ou sacar. No tênis de competição, é comum que o jogador que serve fature o game, já que tem a vantagem do ataque e dita o ritmo do jogo. Desta forma, uma das estratégias de jogo é tentar inverter esta vantagem durante a troca de bola ou durante a defesa fazer com que o adversário, através de erros, perca os games em que está sacando. Ganha o jogobr/encontropt aquele que atingir um número de sets pré-definido — geralmente 2 sets, sendo de 3 sets para os grandes torneios masculinos."@pt .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/ontology/abstract> "Il tennis, il cui nome può essere tradotto in italiano con un termine arcaico non più in uso di “schiaffo palla” e storicamente noto col nome di \"pallacorda\", è uno sport della racchetta che vede opporsi due o quattro giocatori (due contro due, incontro di doppio) in un campo da gioco diviso in due metà da una rete alta 0,914 m al centro e 1,07 m ai lati. È considerato uno sport completo e armonico (sebbene fisicamente in parte asimmetrico perché un braccio viene escluso dal movimento del corpo) in quanto sono richieste al contempo ottime capacità fisiche (coordinazione, velocità e resistenza sulla distanza) e mentali (tecnica nei colpi, tattica, visione di gioco, riflessi e intuito)."@it .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/ontology/abstract> "Tenis (angl. tennis < angl. tenes, tenetz < fr. tenez! = berte, držte! (imperativ pl. slova držet)), označovaný také jako bílý sport, je míčová hra pro 2 nebo 4 hráče. Varianta se 2 hráči se nazývá , varianta se 4 hráči pak čtyřhra. Hraje se také smíšená čtyřhra, při které v každé dvojici hraje jedna žena a jeden muž. Tenisový míč Soupeři stojí proti sobě, na obdélníkovém hřišti (tenisovém dvorci) a pokoušejí se odrazit tenisový míček tenisovou raketou do pole tak, aby jej soupeř nemohl vrátit nebo aby se s míčkem trefil vedle tenisového dvorce (do autu)."@cs .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/ontology/abstract> "كرة المضرب (أو التنس الأرضي في الترجمات الحرفية) نوع من رياضات الراح والتي يتنافس فيها لاعبان في مباريات فردية، أو فريقان مكونان من لاعبين في مباريات زوجية. كلٌ منهم يحمل مضربا ليستخدمه في ضرب الكرة فوق الشبكة نحو منطقة الخصم. وعدد الضربات ليس محددا، إنما النتيجة تحدد الرابح. كرة المضرب أو التنس هو رياضة أولمبية تُلعب على جميع مستويات المجتمع وفئاته ولجميع الأعمار، يمكن لأي شخص أن يمسك بمضرب ويضرب الكرة بما في ذلك ذوو الاحتياجات الخاصة ومستخدمو الكراسي المتحركة. نشأت لعبة التنس الحديثة في برمنغهام بإنجلترا في أواخر القرن التاسع عشر كرياضة تلعب على العشب، لذا فهي وثيقة الصلة بكل من الألعاب الميدانية كالكريكت والبولينغ، إضافة إلى رياضة المضرب القديمة. تغيرت قواعد كرة المضرب الحديثة قليلاً منذ تسعينيات القرن التاسع عشر، باستثناء قاعدتين: من 1908 إلى 1961 كان على اللاعب أن يضع قدمًا واحدة على الأرض في جميع أوقات المباراة، وفي السبعينيات اعتمد الشوط الفاصل (تاي بريك). ومن الإضافات الحديثة إلى اللعبة على المستوى الاحترافي اعتماد تقنية المراجعة الإلكترونية (عين الصقر) إلى جانب نظام تحدي النقاط . يلعب كرة المضرب الملايين من لاعبين، وهي رياضة مشهورة ومنتشرة في جميع أنحاء العالم، وتحظى البطولات الكبرى لكرة المضرب بمتابعة جماهيرية وإعلامية كبيرة، تضعها في مرتبة متقدمة بين الأحداث الرياضية الأكثر أهمية ومتابعة في العالم سنويا. وأهم هذه البطولات: رولان غاروس أو بطولة فرنسا المفتوحة على الملاعب الرملية، بطولة أستراليا المفتوحة على الملاعب الصلبة، بطولة ويمبلدون على الملاعب العشبية، وبطولة الولايات المتحدة المفتوحة على الملاعب الصلبة."@ar .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/ontology/abstract> "Teniso estas rakedsporto, kutime ludata de du homoj, sed ankaŭ foje de kvar homoj. Specifa para ludo estas la ambaŭseksa para ludo, kiam ĉiu teamo konsistas el unu viro kaj unu virino. Estas principe tri kategorioj de tenisejo, nome la gazona, malmola kaj batita terenoj. Dumlude, la ludantoj ĉiam devas revenigi la pilkon per siaj rakedoj al kontraŭa kampo, provante samtempe malebligi tion al oponanto(j)."@eo .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/ontology/abstract> "Tenisa Ingalaterran eta XIX. mendean sortutako kirola da, pilota batekin eta bi edo lau jokalarien artean jokatua. Tenis jokalariei tenislari ere deritze. Mende haren amaieran arautu zuten: Sare batek banatzen duen zelai batean jokatzen da, jokalariak erraketez baliatzen dira pilota sarearen gainetik aurkariaren zelaira botatzeko, partidek ez dute iraupen jakinik (beharrezko puntuazioa lortzean bukatzen da), prestakuntza fisikoa eta teknika behar dira tenisaren ezaugarriak eta arautegia betetzeko."@eu .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/ontology/abstract> "Tenis atau bola tampel adalah olahraga yang biasanya dimainkan antara dua pemain atau antara dua pasangan masing-masing dua pemain. Setiap pemain menggunakan raket untuk memukul bola karet. Tujuan permainan adalah memainkan bola dengan cara tertentu sehingga pemain lawan tidak dapat mengembalikan bola tersebut. Tenis adalah salah satu cabang olahraga Olimpiade dan dimainkan pada semua tingkat masyarakat di segala usia. Olahraga ini dapat dimainkan oleh siapa saja, termasuk orang-orang yang menggunakan kursi roda. Permainan tenis modern berasal dari Birmingham, Inggris pada akhir abad ke-19 sebagai \"tenis lapangan rumput\". Peraturan tenis berubah sedikit sejak 1890-an. Dua perubahan kecil adalah sejak 1908 hingga 1961 pemain yang melakukan service (pukulan pertama) harus menjaga salah satu kakinya tetap di tanah hingga service berpindah dan adopsi sistem tie-break pada 1970-an. Tambahan terakhir yang diterapkan pada tenis profesional adalah teknologi tinjauan ulang elektronik. Tenis dimainkan oleh jutaan orang sebagai olahraga rekreasi dan juga merupakan olahraga tontontan populer di seluruh dunia.Empat kejuaraan tenis terkemuka adalah Australia Terbuka yang dimainkan di lapangan keras, Prancis Terbuka yang dimainkan di lapangan tanah liat, Wimbledon yang dimainkan di lapangan rumput, dan AS Terbuka yang dimainkan juga di lapangan keras."@in .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/property/venue> <http://dbpedia.org/resource/Tennis_court> .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/property/equipment> <http://dbpedia.org/resource/Net_sport> .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/property/equipment> <http://dbpedia.org/resource/Tennis_ball> .
<http://dbpedia.org/resource/Tennis> <http://dbpedia.org/property/equipment> <http://dbpedia.org/resource/Racket_(sports_equipment)> .