A Map of the Stack

Hi I'm Peadar Coyle
(Pronounced PAH-DER - I'm Irish)!!
@springcoil
All views are my own and do not represent any future, current or past employers.

Who I've worked with


Contributor to PyMC3 and other open source software
Author and Speaker at PyData and EuroSciPy
Check out 'Interviews with Data Scientists' - 24 data scientists interviewed - proceeds go to NumFOCUS
My current Adventure
Very excited to be joining Channel 4 in London from early April to work on customer segmentation and recommendation engines
Channel 4 is an award winning not-for-profit TV channel and digital channel.


Version 3 is the way forward!
It's March 2016 I want to do Analytics in PyData
- It depends what you want to do
- This talk includes sample code
- What is new, what is not new, etc
- Very influenced by Rob Story
- I'll talk a bit more about Statistics and ML
- There'll be no bikes in my talk :)
Why use Python for Analytics anyway?
- Although Python is not very fast for things like webservers (Go would be better) it is **very** fast for things like HPC or Numerics.
- Because of C and Fortran (and maybe others like Rust or Theano in the future)
- (HT: Rob Story)
PyData strengths
- A rich and varied ecosystem
- Lots of activities, lots of different ways of attacking the same problems
- A great history of attacking hard problems by community effort

And many others.
Open Source can't thrive without industrial and academic support
Thanks to these guys and girls...




The PyData Stack
And many many more...









Our wonderful ecosystem
- I'll talk about what is new in PyData
- I'll talk about what tools to use with different localities of datasets (in memory, out-of-core and distributed)
- I'll talk about our Statistical tools including PyMC3
- I'll talk about the future!!
What's new and useful in PyData
New in the core of the stack
-
It is impossible to talk about PyData without mentioning NumPy and Pandas

Improvements throughout the stack
Matplotlib colours, Sympy new release, improvements in NumPy
New @ operator in NumPy
Assign, and pipe in Pandas
Pandas - assign
df = pd.DataFrame({'A': range(1, 11), 'B': np.random.randn(10)})
df.assign(ln_A_plus_1=lambda x: np.log(x.A)+1)Creates a copy of the dataframe with a nice new column.
Really useful for percentages, logarithms etc - standard Financial Analysis and Data Analysis stuff.

I have a data problem to solve
In practice grouping and counting things :)
Adult data set
data
age workclass fnlwgt education-categorical educ marital-status
0 39 State-gov 77516 Bachelors 13 Never-married
2 38 Private 215646 HS-grad 9 Divorced
3 53 Private 234721 11th 7 Married-civ-spouse
4 28 Private 338409 Bachelors 13 Married-civ-spouse
5 37 Private 284582 Masters 14 Married-civ-spouse
6 49 Private 160187 9th 5 Married-spouse-absent Source UCI Adult data set, csv version here: http://pymc-devs.github.io/pymc3/Bayesian_LogReg/
I can only use standard library
I'm stuck on a restricted machine and I only have Python 2.6
(Example shamelessly stolen from Rob Story and adapted for my data set)
import csv
conversion_map = {
'age': int,
'workclass': str,
'fnlwgt': int,
'education-categorical': str,
'educ': int,
'occupation': str,
'sex': str,
'capital-gain': float,
'capital-loss': float,
'hours': int,
'native-country': str,
'income': str
}
Write a conversion map and use csv
Load the csv data source
def converter(type_map, row):
"""Yep, we need to roll our own type conversions."""
converted_row = {}
for col, val in row.items():
converter = type_map.get(col)
if converter:
converted_row[col] = converter(val)
else:
converted_row[col] = val
return converted_row
with open('adult.csv', 'r') as f:
reader = csv.DictReader(f)
adult2 = [converter(conversion_map, r) for r in reader]How does it look
>>> adult2[:2]
[{'': '0',
'age': 39,
'capital-loss': 0.0,
'captial-gain': '2174',
'educ': 13,
'education-categorical': ' Bachelors',
'fnlwgt': 77516,
'hours': 40,
'income': ' <=50K',
'marital-status': ' Never-married',
'native-country': ' United-States',
'occupation': ' Adm-clerical',
'relationship': ' Not-in-family',
'sex': ' Male',
'workclass': ' State-gov'},I want to get the maximum age in my dataset
def get_max_age():
max_age = 0
for row in adult2:
if row['age'] > 1 and row['age'] > max_age:
max_age = row['age']
return max_age
>>> get_max_age()
90
# Or you could do it like this generator expression
>>> max(row['age'] for row in adult2 if row['age'] > 1)
90Let's say you wanted to group things
# defaultdict is awesome. defaultdict is awesome.
from collections import defaultdict
def grouper(grouping_col, seq):
"""People have definitely written a faster version than what I'm about to write
Thanks to Rob Story for this one"""
groups = defaultdict(lambda: defaultdict(list))
for row in seq:
group = groups[row[grouping_col]]
for k, v in row.items():
if k != grouping_col:
group[k].append(v)
return groups
>>> groups = grouper('occupation', adult2)A natural question is the mean number of hours by occupation
summary = {}
for group, values in groups.items():
summary[group] = sum(values['hours']) / len(values['hours'])
>>> summary
{' ?': 31.90613130765057,
' Adm-clerical': 37.55835543766578,
' Armed-Forces': 40.666666666666664,
' Craft-repair': 42.30422054159551,
' Exec-managerial': 44.9877029021151,
' Farming-fishing': 46.989939637826964,
' Handlers-cleaners': 37.947445255474456,
' Machine-op-inspct': 40.755744255744254,
' Other-service': 34.70166919575114,
' Priv-house-serv': 32.88590604026846,
' Prof-specialty': 42.38671497584541,
' Protective-serv': 42.87057010785824,
' Sales': 40.78109589041096,
' Tech-support': 39.432112068965516,
' Transport-moving': 44.65623043206011}Interlude: Itertools
It is common advice but it's worth being aware of itertools if you want to write something like this.
I wanna count things - in a functional way
PyToolz
PSA: PyToolz is awesome allows you to use functional programming techniques in Python.
http://toolz.readthedocs.org/en/latest/index.html
I want to make it faster - I'll use CyToolz
PyToolz example
#I wanna see the frequencies of ages in the dataset
>>> tz.frequencies([r['age'] for r in adult2])
# Toolz has currying!
#I want to count by all of the occupations with greater than 15 years of education
import toolz.curried as tzc
>>> tzc.pipe(adult2,
tzc.filter(lambda r: r['educ'] > 15),
tzc.map(lambda r: (r['occupation'],)),
tzc.countby(lambda r: r[0]),
dict)
{' ?': 15,
' Adm-clerical': 5,
' Craft-repair': 2,
' Exec-managerial': 55,
' Farming-fishing': 1,
' Machine-op-inspct': 1,
' Other-service': 1,
' Prof-specialty': 321,
' Sales': 8,
' Tech-support': 3,
' Transport-moving': 1}Summary: Toolz
Toolz has some great virtues
-
Composability: They interoperate due to core data structures
-
Purity: They don't change their input or rely on external state
-
Lazy: Only evaluated when needed
- They also support serializability so they're easy to accelerate or parallelize
Interlude
Pandas
Not going to talk too much about Pandas in this talk.
It is fast becoming a stable and core member of the PyData stack
Really useful for indexed data like time series data or csv file data
Statsmodels and seaborn already consider it a core member of the stack
- I won't talk about Numpy either
- Xarray and Dask are all either dependent on it or strongly influenced by it
- Pandas depends on it
- Many other projects like Scipy depend on it
- The speed optimizations and the ability to release the GIL allow this to be very fast for modern hardware
- Recent improvements include the '@' operator making it a lot easier to write good linear algebra code in NumPy

End of Interlude

Labelled heterogenous data
NumPy arrays plus labels - excellent for 'Scientific data' :)
I have weather forecasting data in NetCDF - this is what you use
Xarray looks like this
arr = np.array([[1, 2, 3, 4],
[10, 20, 30, 40],
[100, 200, 300, 400]])
dim0_coords = ['a', 'b', 'c']
dim1_coords = ['foo', 'bar', 'baz', 'qux']
da = xray.DataArray(arr, [('x', dim0_coords), ('y', dim1_coords)])
da
da.loc['b']There are plenty of examples in the notebooks
>> da[0:3]
<xarray.DataArray (x: 3, y: 4)>
array([[ 1, 2, 3, 4],
[ 10, 20, 30, 40],
[100, 200, 300, 400]])
Coordinates:
* x (x) <U1 'a' 'b' 'c'
* y (y) <U3 'foo' 'bar' 'baz' 'qux'
>>> da.dims
('x', 'y')
>> da.coords
Coordinates:
* x (x) <U1 'a' 'b' 'c'
* y (y) <U3 'foo' 'bar' 'baz' 'qux'
# Get a mean by label
>> da.mean(dim='y')
<xarray.DataArray (x: 3)>
array([ 2.5, 25. , 250. ])
Coordinates:
* x (x) <U1 'a' 'b' 'c'- I want to disconnect or 'decouple' my expressions for computations from my backend
- Why can't I do Pandas like things on Postgresql?
- I have some data in CSV, some in HDF5 (PyTables), some in my SQL database
- I still run into in-memory problems
- I have bigger-than-I-can-RAM-data
- Getting a bigger machine is overkill
- Why are my analytical expressions tied to my data structure?
- Can I have expressions that work across data structure *and* storage?

-
Blaze: An interface to query data on different storage systems
-
Dask: Parallel computing through task scheduling and blocked algorithms
-
Datashape: A data description language
- DyND: A C++ library for dynamic, multidimensional arrays
- Odo: Data migration between different storage systems
Blaze Ecosystem
We'll talk about Dask later
- We'll use Odo and Blaze in this demo
- Datashape and DyND are awesome but I won't talk about them
import blaze as bz
bz_adult = bz.symbol('adult2', bz.discover(adult))
>>> type(bz_adult)
blaze.expr.expressions.Symbol
>>> mean_age = bz.by(bz_adult.occupation,
price=bz_adult.age.mean())
>>> hours_count = bz.by(bz_adult[bz_adult.hours > 35].educ,
count=bz_adult.workclass.count())
# We haven't actually computed anything yet!
# Let's make Pandas compute it.
bz.compute(mean_age, adult)# We have here the count of number of years of education
# by a certain filter of greater than 35 hours of work per week.
>>> bz.compute(hours_count, adult)
educ count
0 1 51
1 2 168
2 3 333
3 4 646
4 5 514
5 6 933
6 7 1175
7 8 433Let's compute in Postgres!
# Blaze/Odo make it easy to move data between containers
# Note that we have an empty table already created
pg_datasource = bz.odo(adult,
"postgresql://peadarcoyle@localhost/pydata::adult2")
# Now we're going to use Postgres as our computation engine
result = bz.compute(hours_count, pg_datasource)
result
<sqlalchemy.sql.selectable.Select at 0x113ae4390; Select object>
# I don't want a selectable. I want a DataFrame
# odo again
bz.odo(bz.compute(hours_count, pg_datasource), pd.DataFrame)
educ count
0 8 433
1 16 413
2 15 576
3 4 646
4 1 51Let's store in Bcolz (we'll see Bcolz and ctable- the storage format later)
import bcolz
>> %time bz.odo(adult, 'adult.bcolz')
CPU times: user 10.3 s, sys: 18.1 s, total: 28.4 s
Wall time: 28.8 s
Out[55]:
ctable((32561,), [('age', '<i8'), ('workclass', 'O'), ('fnlwgt', '<i8'),
('educationcategorical', 'O'), ('educ', '<i8'), ('maritalstatus', 'O'),
('occupation', 'O'), ('relationship', 'O'), ('sex', 'O'), ('captialgain', '<i8'),
('capitalloss', '<i8'), ('hours', '<i8'), ('nativecountry', 'O'), ('income', 'O')])
nbytes: 7.76 MB; cbytes: 43.54 MB; ratio: 0.18
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
rootdir := 'adult.bcolz'
[ (39, ' State-gov', 77516, ' Bachelors', 13, ' Never-married', ' Adm-clerical',
' Not-in-family', ' Male', 2174, 0, 40, ' United-States', ' <=50K')
(50, ' Self-emp-not-inc', 83311, ' Bachelors', 13, ' Married-civ-spouse',
' Exec-managerial', ' Husband', ' Male', 0, 0, 13, ' United-States', ' <=50K')
(38, ' Private', 215646, ' HS-grad', 9, ' Divorced', ' Handlers-cleaners',
' Not-in-family', ' Male', 0, 0, 40, ' United-States', ' <=50K')
...,
(58, ' Private', 151910, ' HS-grad', 9, ' Widowed', ' Adm-clerical',
' Unmarried', ' Female', 0, 0, 40, ' United-States', ' <=50K')
(22, ' Private', 201490, ' HS-grad', 9, ' Never-married', ' Adm-clerical',
' Own-child', ' Male', 0, 0, 20, ' United-States', ' <=50K')
(52, ' Self-emp-inc', 287927, ' HS-grad', 9, ' Married-civ-spouse',
' Exec-managerial', ' Wife', ' Female', 15024, 0, 40, ' United-States', ' >50K')]What else?
You can use any SQL supported by SQLAlchemy as your computation. It also supports Python lists, Spark DataFrames, MongoDB, Numpy arrays...
I want to maximize my speed of reading/writing on a single computer
-
bcolz is a columnar data store for fast data storage and retrieval with built-in high performance compression. It supports both in-memory and out-of-memory storage and operations. Cf. http://bcolz.blosc.org/.
Bcolz
- Fast IO and leverages Blosc for compression
- For certain problems like reading timeseries and doing analytics this can be useful
- We've seen immature projects like Castra built on top of it.
- We'll see more and more tools leveraging fast compression structures.
- Here I use POIWorld a dataset of 'Points of Interest' from OpenStreetMap
- Has some great synergies with binary formats like HDF5

df_poiworld = pd.read_csv('POIWorld.csv', usecols=columns)
dc = bcolz.ctable.fromdataframe(df_poiworld)
dc
ctable((9140052,), [('name', 'O'), ('amenity', 'O'),
('Longitude', '<f8'), ('Latitude', '<f8')])
nbytes: 575.61 MB; cbytes: 3.00 GB; ratio: 0.19
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
[(nan, 'post_box', -0.20698000000000003, 51.9458753)
(nan, 'post_box', -0.268633, 51.938183)
(nan, 'post_box', -0.274278, 51.930209999999995) ...,
(nan, nan, -77.2697855, 39.24023820000001)
(nan, nan, -77.2777191, 39.237238399999995)
(nan, 'drinking_water', -5.8, nan)]>>> dc.cols
age : carray((32561,), int64)
nbytes: 254.38 KB; cbytes: 256.00 KB; ratio: 0.99
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
[39 50 38 ..., 58 22 52]
workclass : carray((32561,), |S17)
nbytes: 540.56 KB; cbytes: 303.83 KB; ratio: 1.78
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
[b' State-gov' b' Self-emp-not-inc' b' Private' ..., b' Private'
b' Private' b' Self-emp-inc']
educ : carray((32561,), int64)
nbytes: 254.38 KB; cbytes: 256.00 KB; ratio: 0.99
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
[13 13 9 ..., 9 9 9]
occupation : carray((32561,), |S18)
nbytes: 572.36 KB; cbytes: 338.49 KB; ratio: 1.69
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
[b' Adm-clerical' b' Exec-managerial' b' Handlers-cleaners' ...,
b' Adm-clerical' b' Adm-clerical' b' Exec-managerial']
sex : carray((32561,), |S7)
nbytes: 222.58 KB; cbytes: 256.00 KB; ratio: 0.87
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
[b' Male' b' Male' b' Male' ..., b' Female' b' Male' b' Female']
hours : carray((32561,), int64)
nbytes: 254.38 KB; cbytes: 256.00 KB; ratio: 0.99
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
[40 13 40 ..., 40 20 40]%%time
#Generate 1GB of data
>> N = 100000 * 1000
>> import bcolz
>> ct = bcolz.fromiter(((i, i ** 2) for i in range(N)),
dtype="i4, i8",
count=N,
cparams=bcolz.cparams(clevel=9))
CPU times: user 59.6 s, sys: 1.08 s, total: 1min
Wall time: 59.1 s
>> ct
ctable((100000000,), [('f0', '<i4'), ('f1', '<i8')])
nbytes: 1.12 GB; cbytes: 151.84 MB; ratio: 7.54
cparams := cparams(clevel=9, shuffle=True, cname='blosclz')
[(0, 0) (1, 1) (2, 4) ..., (99999997, 9999999400000009)
(99999998, 9999999600000004) (99999999, 9999999800000001
That is 7 times compression in-memory
You can also store on disk and read it fast
>> %time ct.eval('f0 ** 2 + sqrt(f1)')
CPU times: user 4.38 s, sys: 1.96 s, total: 6.34 s
Wall time: 1.26 s
Out[36]:
carray((100000000,), float64)
nbytes: 762.94 MB; cbytes: 347.33 MB; ratio: 2.20
cparams := cparams(clevel=5, shuffle=True, cname='blosclz')
[ 0.00000000e+00 2.00000000e+00 6.00000000e+00 ..., 1.37491943e+09
1.57491943e+09 1.77491942e+09]Fast numerical calculations
Integration with Numexpr to handle expressions
Intelligent use of caching and multithreading to optimize numerical calcuations
Let's look at Adult dataset again
- With Bcolz you can do Pandas like things
- Based on NumPy but has support for PyTables/HDF5 (which may be faster)
- Uses chunking. The chunked nature of bcolz objects, together with buffered I/O, makes appends very cheap
- This makes this ideal for say storing and retrieving market data.
- This is for fast fetch, and write rarely...
dc['workclass' == ' State-gov']
#dc.cols
# You can do DataFrame-like stuff
dc['workclass' == ' State-gov']
Out[117]:
(39, b' State-gov', 13, b' Adm-clerical', b' Male', 40)PSA: Bcolz version 1 release candidate is out
There are some challenges with integration into the rest of PyData, this should stabilize.
Quantopian Inc a crowd-sourced hedge fund uses Bcolz
- Each Column Is Stored Separately
- Escapes the GIL
- Better compression ratio for binary data
- Allows you to compress in-memory/ on disk and retrieve fast
- https://quantopian.github.io/talks/NeedForSpeed/slides.html

My data is bigger than RAM or in a cluster
Use Dask or specifically dask.array
Dask looks like this!
Basically the Pandas/NumPy API
import dask.array as da
# create a dask array from the above array
a2 = da.from_array(a, chunks=200)
# multiply this array by a factor
b2 = a2 * 4
# find the minimum value
b2_min = b2.min()
print(b2_min)
#I want to tell if this is a School
#or not and then plot it on a graph
>> is_school = with_amenity.amenity.str.contains('[Ss]chool')
>> school = with_amenity[is_school]
#Very similar to pandas but you need to
#call compute on the dask objects
>> dd.compute(school.amenity.count())
(342025,)
# So we have about 342k schools in
# UK and Ireland in the OpenStreetMap projectimport dask.dataframe as dd
lon, lat = dd.compute(school.Longitude,
school.Latitude)
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap
fig, ax = plt.subplots(figsize=(10, 15))
m = Basemap(projection='mill',
lon_0=-5.23636, lat_0=53.866772,
llcrnrlon=-10.65073, llcrnrlat=49.16209,
urcrnrlon=1.76334, urcrnrlat=60.860699)
m.drawmapboundary(fill_color='#ffffff', linewidth=.0)
x, y = m(lon.values, lat.values)
m.scatter(x, y, s=1, marker=',', color="steelblue", alpha=0.6);Compute in Dask and plot in Matplotlib
Notice how similar to Pandas and NumPy the API is.
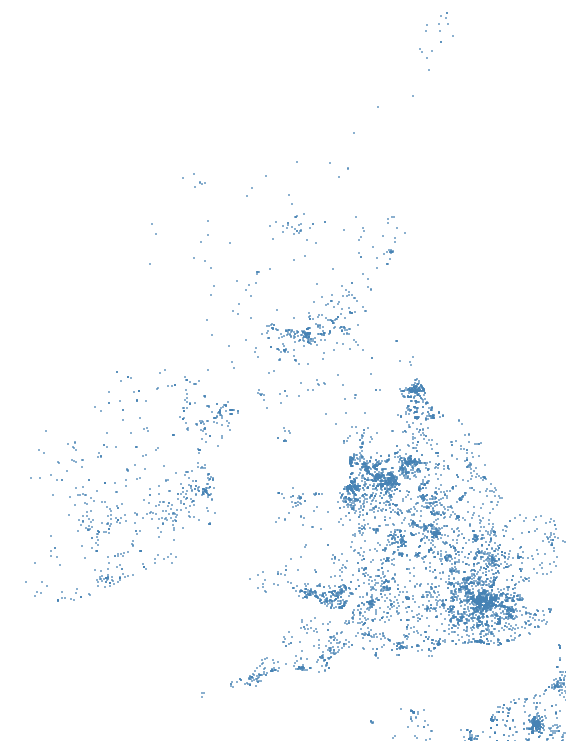
UK and Irish schools in Open Street Map
When do you use Dask?
- Medium data is greater than RAM size
- Generally Dask comes into it's own on around 16GB datasets
- Anything around the 1TB - 1PB range probably needs either a good SQL database or something like Spark
- Benchmark: My own Laptop has 4 cores and about 8GB of RAM
Distributed Arrays
- Bolt - distributed arrays backed by Spark
- DistArray - other distributed arrays
- Biggus - another kind of distributed array (virtual)
- Dask.array - Distributed arrays using task scheduling

Very exciting technology for the JVM community
Improvements in PySpark and interoperability
Improvements in Machine Learning libraries
Comes into it's own with lots of JSON blobs on many nodes
Interlude
I want to speed up my code
- Numba (A fast LLVM based JIT compiler that is easy to use via decorators)
- Cython (A language that allows you to blend C objects for great speedup)
- PyPy (Another compiler but without support for NumPy code) Recent improvements in PyPy
- Plenty of tutorials online, and new tools are arriving...
End of Interlude
Recent improvements in dealing with 'Big Data'
- Distributed computing has improved in Dask
- See Matt Rocklins website
- PyData will just get better and better at dealing with 'big data'
- Soon you may not need to use the JVM to deal with HDFS.
- Spark is improving too. Spark is very exciting and I could give an entire talk in Spark. Other people are doing that!
Arrow and Ibis
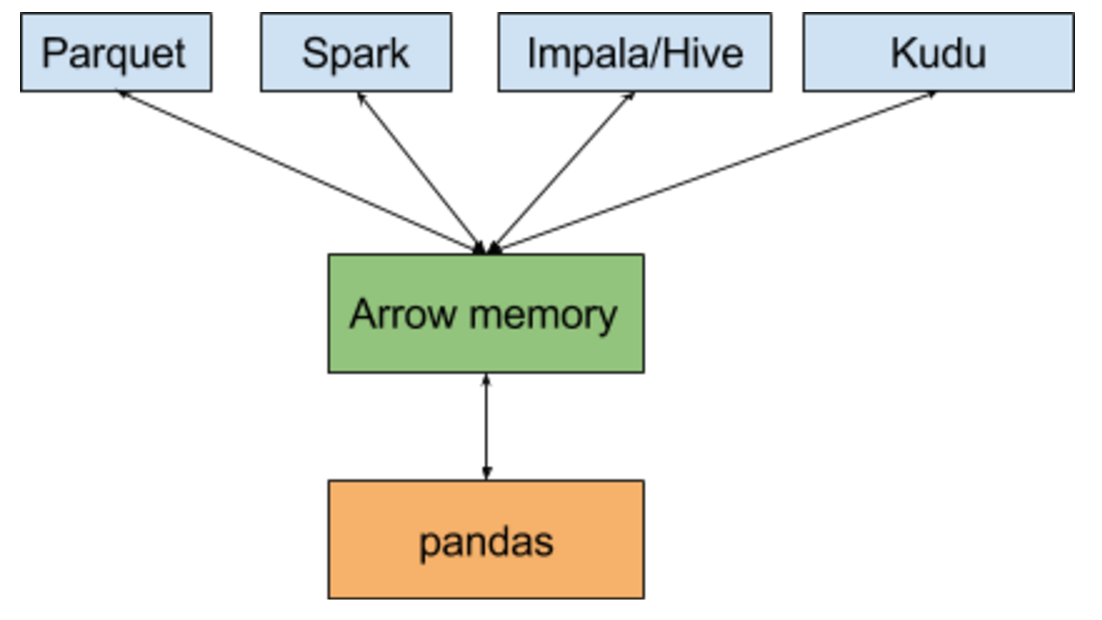
Source: Wes McKinney
Ibis Example
rounds = con.table('pokemon_types')
rounds.info()
#This is a Pokemon table in SQLite
rounds.slot.value_counts()
slot count
0 1 784
1 2 395SQLite in the background but could be Impala - all with a pandas like API
PyData Map
Wouldn't it be great to have a map for the stack
I had a a go
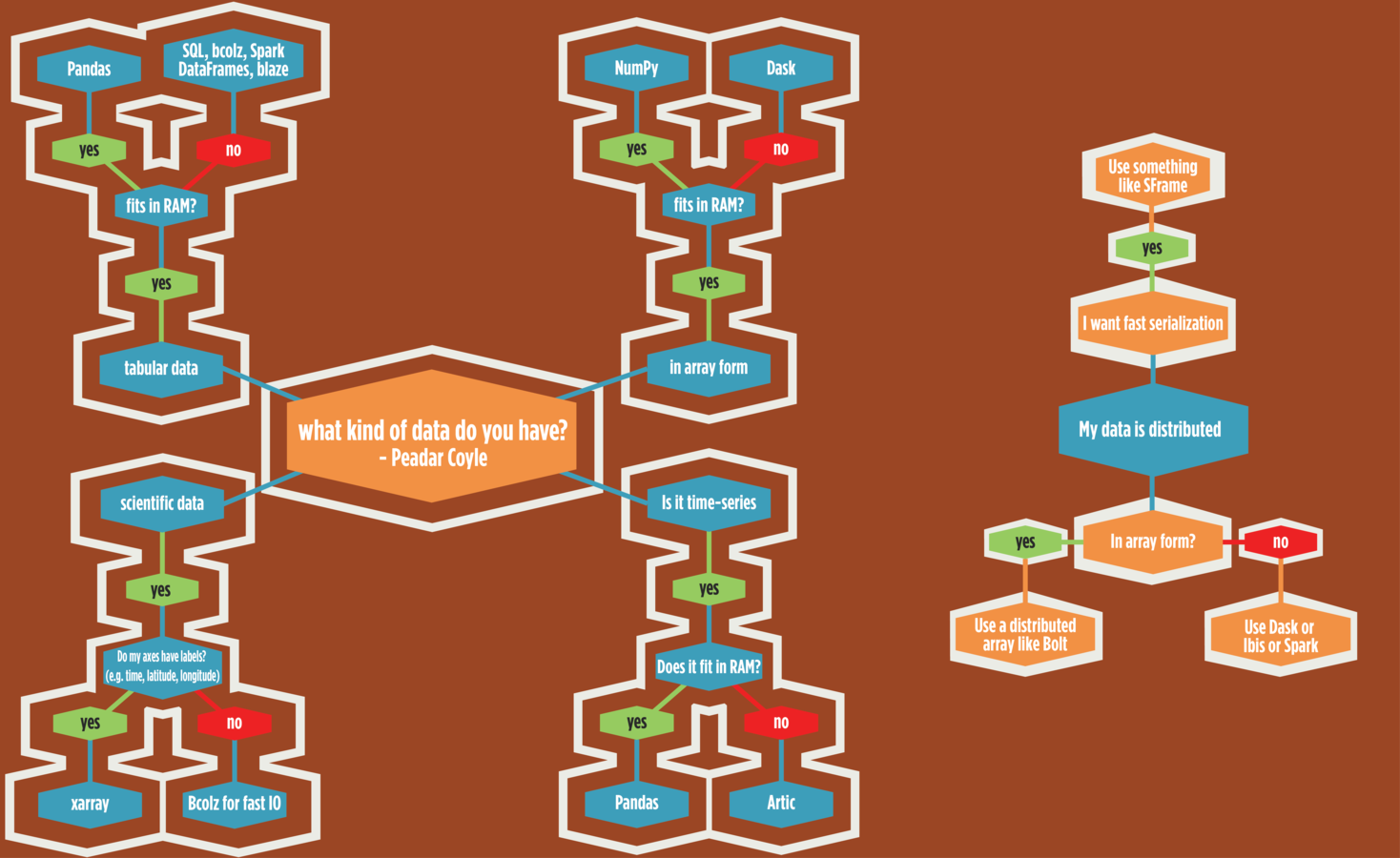
I wanna do Stats/ML
There's lots of cool stuff in PyData Land!
PyMC3
- Recent improvements to documentation!!
- Written on top of Theano
- Timeseries examples, Bayesian Logistic Regression
- Model evaluation functions
- PSA: It's now in Beta

Bayesian LogReg
Subtitle
data[data['native-country']==" United-States"]
income = 1 * (data['income'] == " >50K")
age2 = np.square(data['age'])
data = data[['age', 'educ', 'hours']]
data['age2'] = age2
data['income'] = income
with pm.Model() as logistic_model:
pm.glm.glm('income ~ age + age2 + educ + hours',
data, family=pm.glm.families.Binomial())
trace_logistic_model = pm.sample(2000,
pm.NUTS(), progressbar=True)
Statsmodels
PSA: If you want to help PyData a lot - work on Statsmodels

Scikit-Learn
- The best documentation in PyData
- Lots of cool improvements
- Chat to Andreas about this - he's at PyData Amsterdam

I want to analyze text


Production ready NLP toolkits all under open source
The future...
What is coming next
- Substantially improved data access speeds
- Closer to native performance Python extensions like Apache Spark
- New in-memory analytics functionality for nested/JSON-like data
Theano


Computational toolkits


Lasagne
So cite, send pull requests and/or help NumFOCUS!


Copy of The PyData Map of the Stack
By springcoil
Copy of The PyData Map of the Stack
Keynote from PyData Amsterdam
- 5,778



