


Computational Biology
(BIOSC 1540)
Feb 11, 2025
Lecture 06A
Read mapping
Foundations
Let's have a discussion
I have to
(1) Prepare computationalists for future classes
(2) Introduce non-computationalists to the field
and
Programming
is a crucial skill for anything computational
is a helpful skill for science
What?
Use?
How?
What can we do with comp bio?
How do we use the user-friendly tools?
How do the tools work?
1
10
5
Let's have a discussion
Changes I can make
A. Simpler and fewer Python problems
B. Offer an optional Python recitation either in the evening or weekend
C. "Flipped" classroom where I record lectures/assign readings and use classtime for Python
D. No changes
E. No Python
Announcements
Assignments
Quizzes
CBytes
ATP until the next reward: 653
After today, you should have a better understanding of
How transcriptomics extends beyond genomics
DNA
Genomics allows us to study the blueprint of organisms
DNA sequences are stable across an organism’s lifetime
Questions genomics can answer:
- What genes are present? (e.g., Does a bacterium have antibiotic resistance genes?)
- How are species related? (e.g., Evolutionary trees based on genome sequences.)
- What mutations exist? (e.g., Cancer-causing genetic changes.)
Genomics helps answer key biological questions
Genomics tells us what’s possible for an organism to do but not when or how it does it.
Examples:
- Every cell in your body has the same genome, but a neuron and a liver cell express different genes.
- In cancer, certain genes are turned on or off incorrectly—but genomics alone can’t detect this.
A genomic sequence alone doesn’t tell us what genes are active
DNA is like a book of instructions—just because a gene exists doesn’t mean it’s being used.
Key insight: To understand cellular function, we need to know which genes are active and when.
After today, you should have a better understanding of
How transcriptomics extends beyond genomics
RNA
Transcriptomics: A real-time microscope
Transcriptomics allows us to see precisely what genes are active at a given moment
We can see gene expression changes over time
Allows us to see which annotated genes are actually being used
The transcriptome is constantly changing and captures the cell's response to its environment and internal signals
- Environmental conditions: Cells respond to stress, nutrients, or pathogens by changing gene expression
- Developmental stage: The genes active in an embryo differ from those in an adult
- Cell type: A neuron will have a different gene expression profile than a liver cell
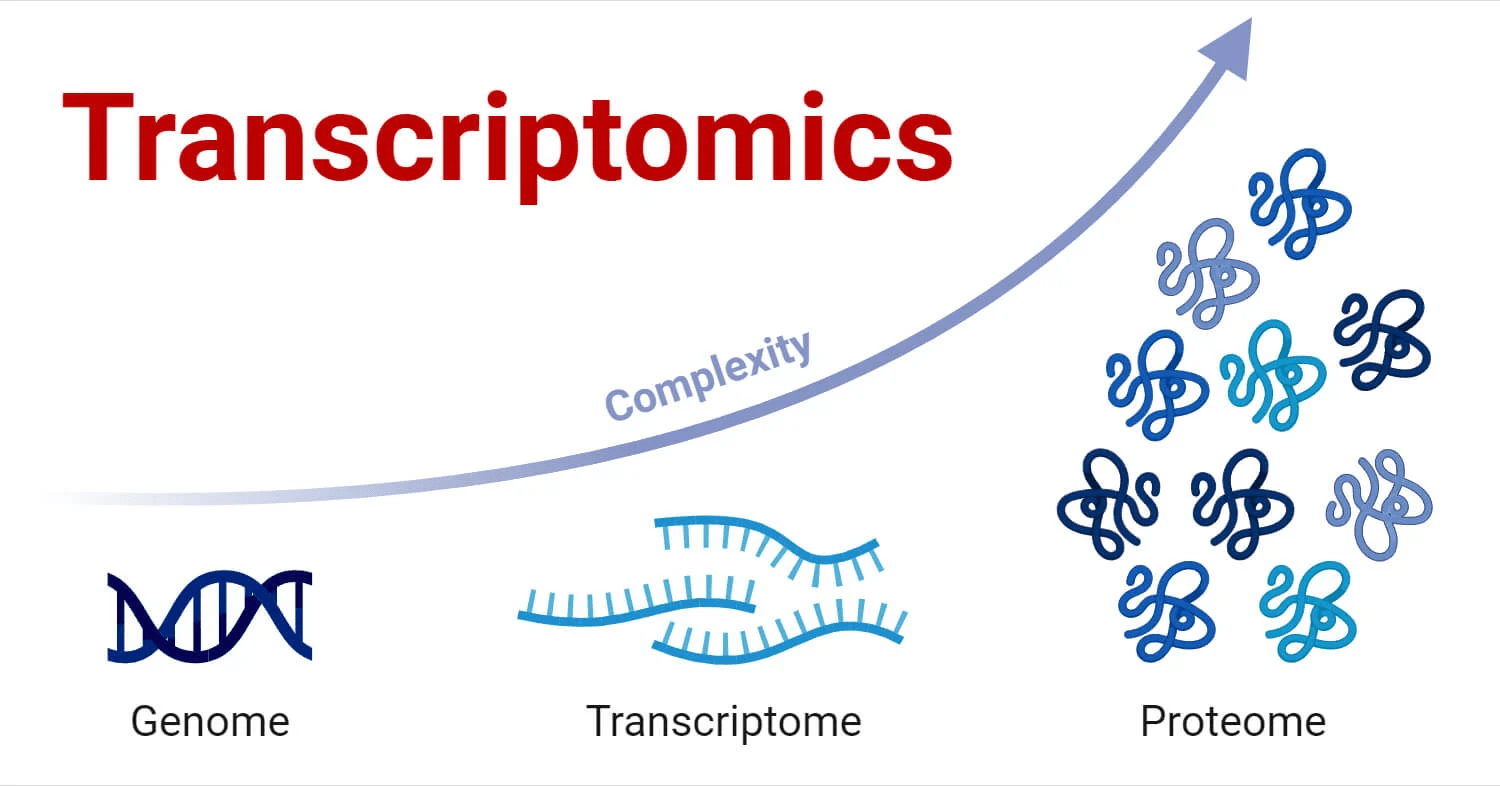
Genomics Provides a Static Blueprint, but Transcriptomics Captures Dynamic Activity

Transcriptomics works with the complete set of RNA transcripts
This includes
mRNA: instructions for protein synthesis
rRNA: forms part of the ribosome structure
tRNA: helps translate the genetic code into proteins
Non-coding RNAs: play regulatory roles in the cell
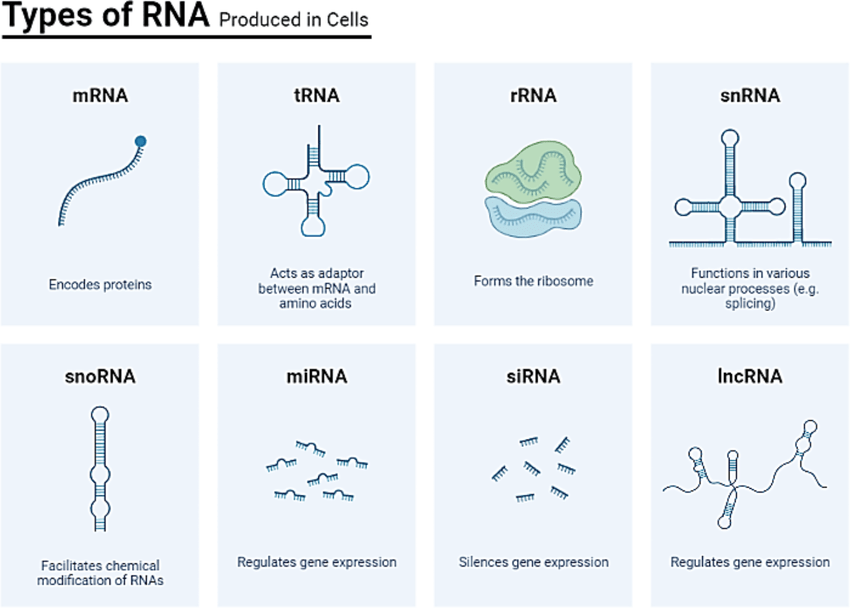



(And more)
Transcriptomics reveals alternative splicing and isoforms
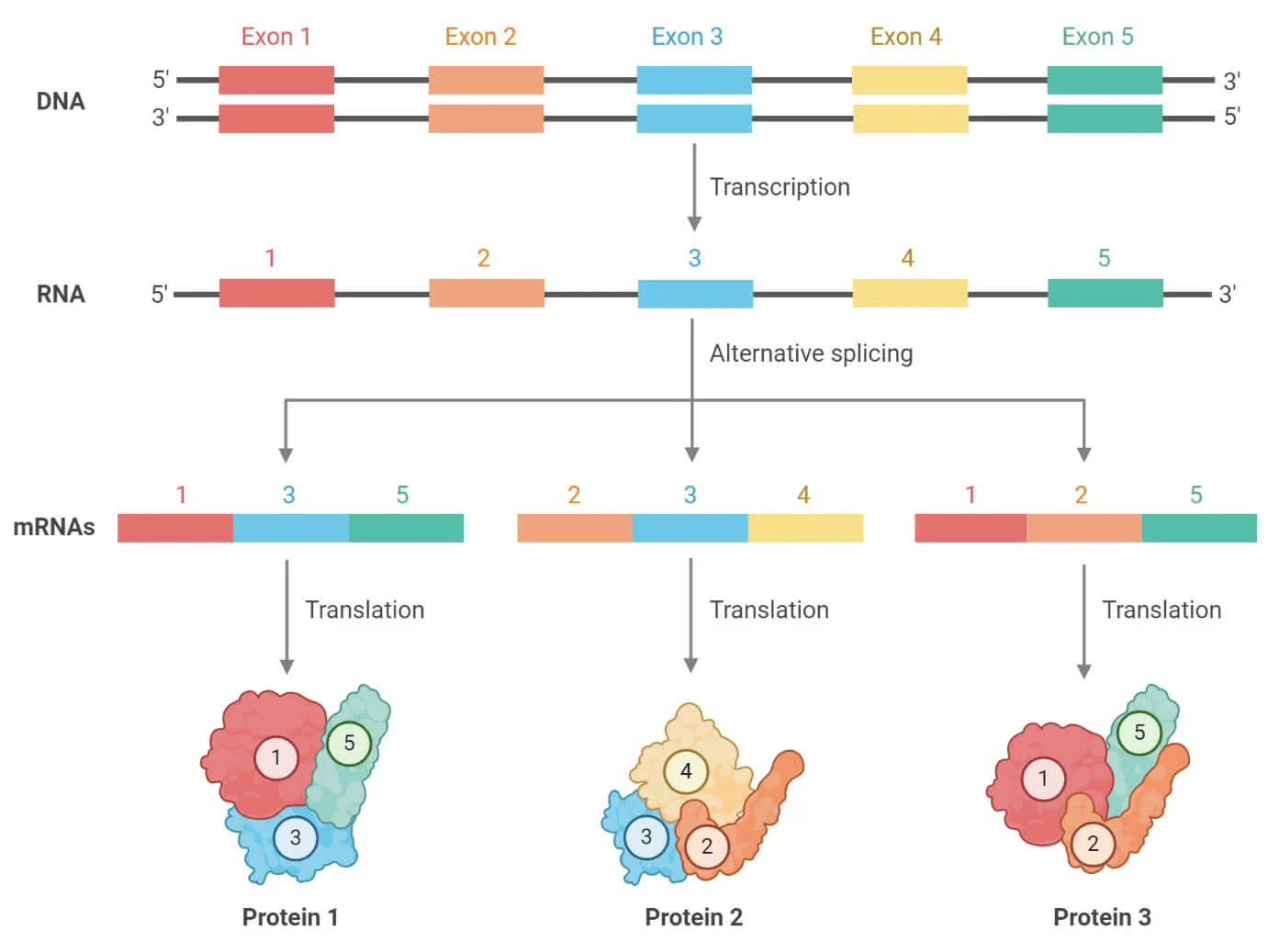
A single gene can produce multiple mRNA transcripts, which we call isoforms
One of the main ways organisms can increase protein diversity without increasing the number of genes
It's estimated that over 90% of human genes undergo alternative splicing
Example: Dscam in Drosophila
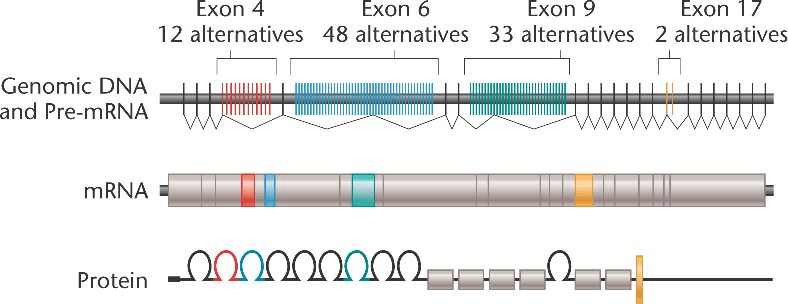
Drosophila melanogaster has over 38,000 isoforms from this one gene
Dscam (Down syndrome cell adhesion molecule) is involved in neural development
Functional insights
- Reveals which elements are active
- Shows diseases state
- Identifies potential functional elements
- Predicts disease risk
Genomics
Transcriptomics
- Requires one-time sampling
- Reveals evolutionary history
- Captures real-time cellular responses
Temporal insights
After today, you should have a better understanding of
The role of RNA-seq in modern transcriptomics
Sample collection
Separate cells from media
Great! We have our cells, but how can we extract our RNA?
The first step is always to centrifuge and separate our cells and media
Keep the part that has our component of interest (RNA)
We break open our cells by lysing them
Chemical lysis destabilizes the lipid bilayer and denatures proteins
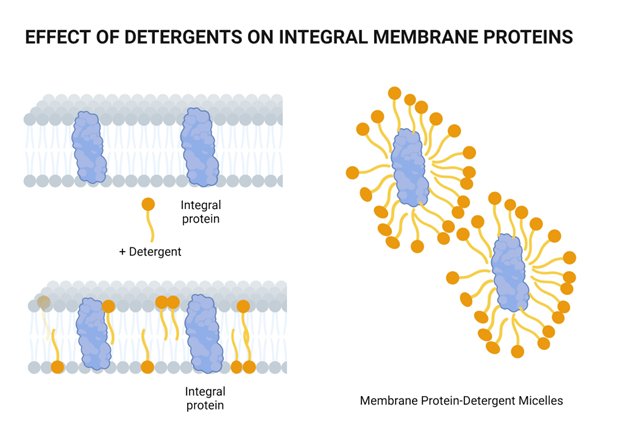
Surfactants have a hydrophilic head and hydrophobic tail
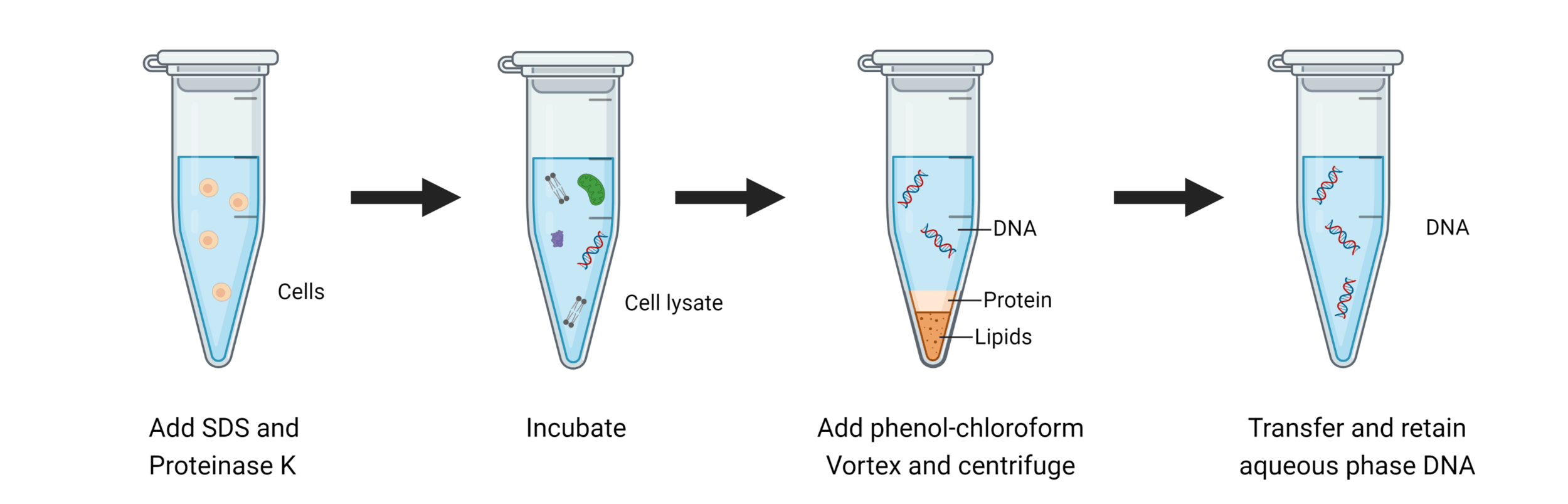
Phenol-chloroform extraction exploits solubility and density differences
Phosphate backbone
(negative charged)
Denatures and aggregates at interface
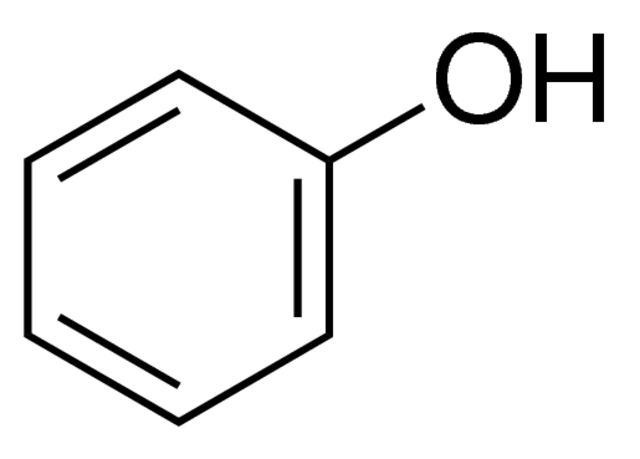
Phenol
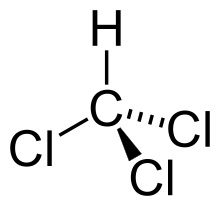
Chloroform
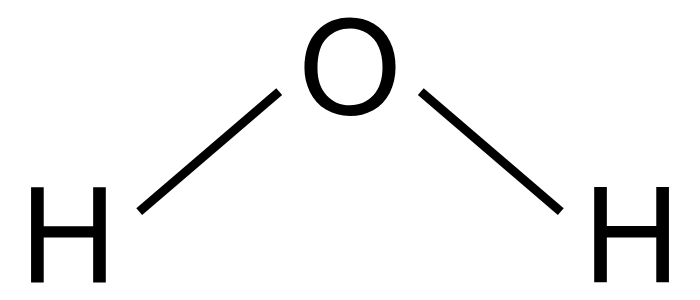
Water
Nonpolar

DNA
RNA

Protein
Lipids
Collecting our aqueous phase selects only DNA and RNA
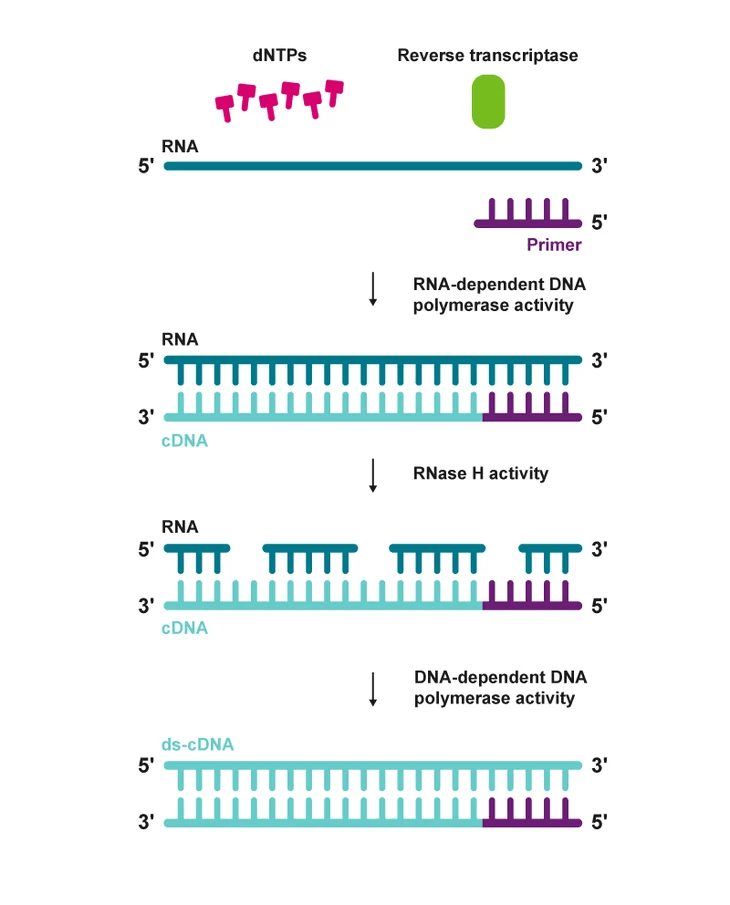
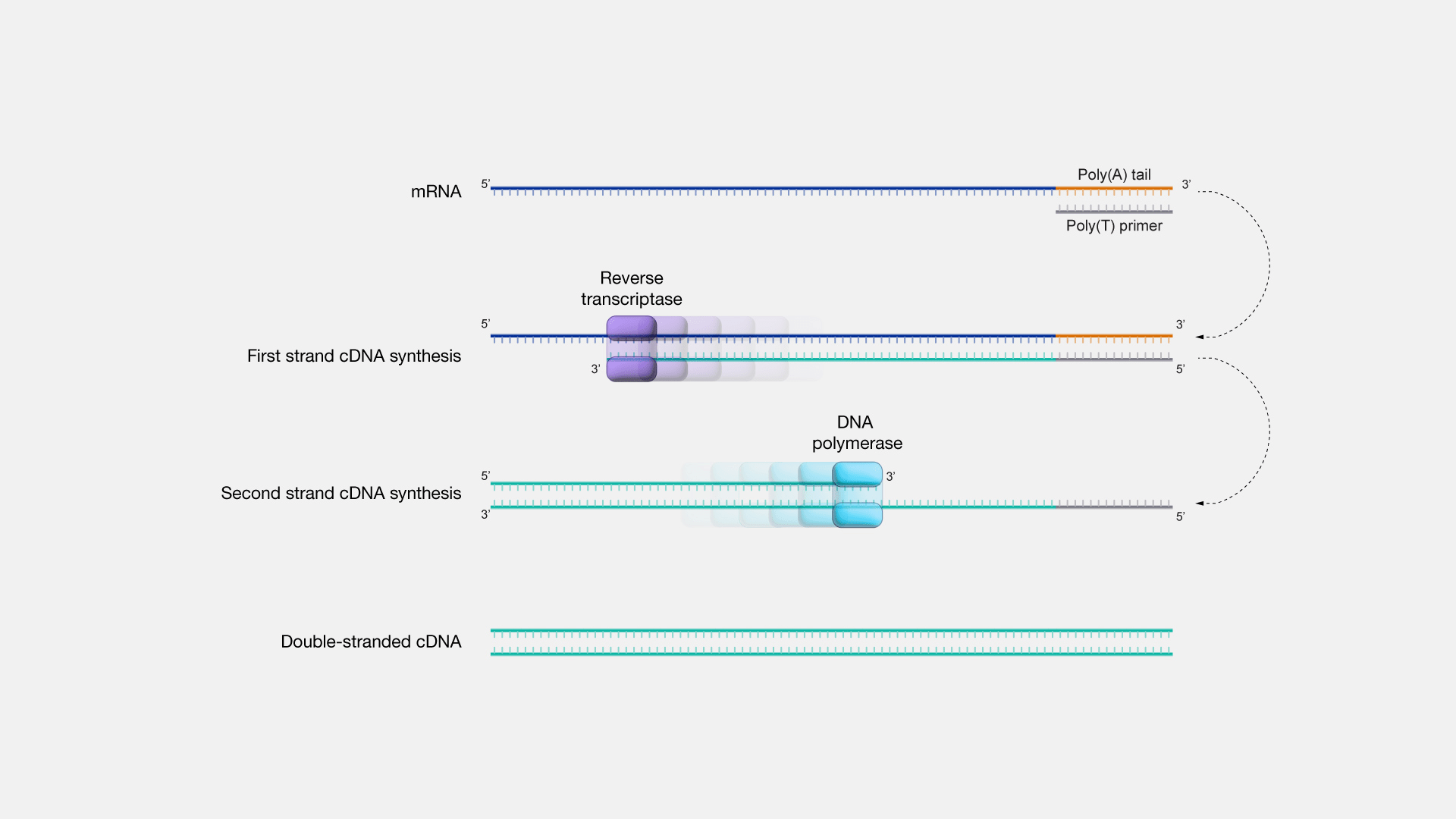
Reverse transcription introduces unique challenges
RNA is converted to cDNA using reverse transcriptase
- Random or oligo(dT) primers influence transcript representation
- Second-strand synthesis method can preserve strand information
mRNA enrichment focuses sequencing on protein-coding transcripts
Enrichment method affects
- Gene expression measurements
- Detection of non-coding RNAs
- Identification of immature transcripts
Poly(A) selection captures mature mRNAs
How could we filter our sample for only mRNA?

After today, you should have a better understanding of
The role of RNA-seq in modern transcriptomics
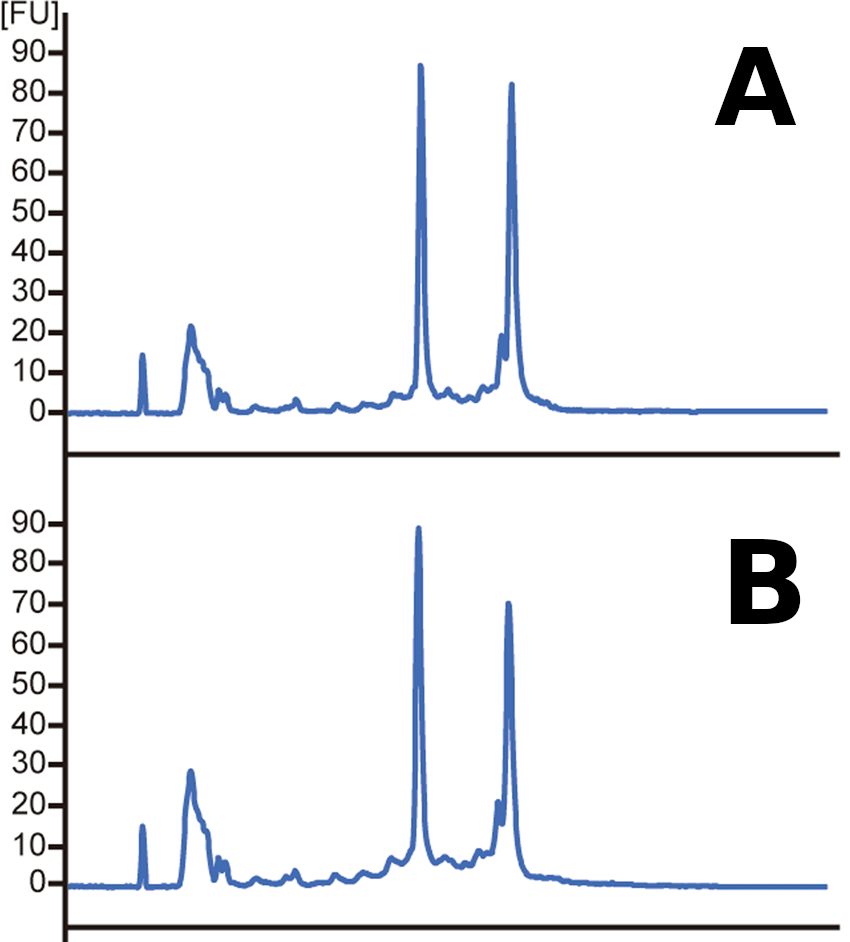
Sample quality
RNase starts degrading RNA rapidly
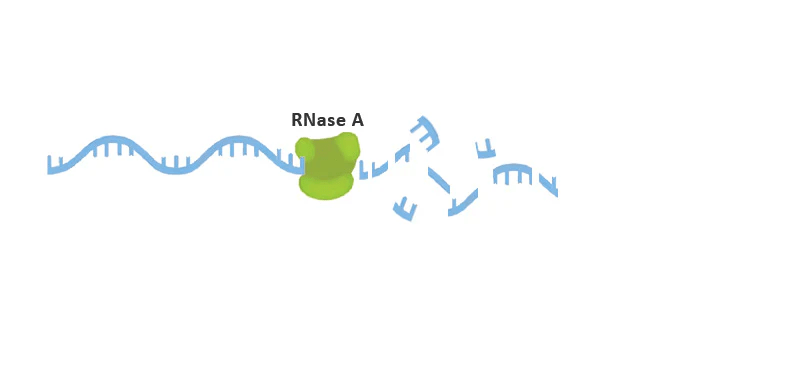
RNA quality is critical for successful sequencing
Assess RNA integrity (RNA Integrity Number)
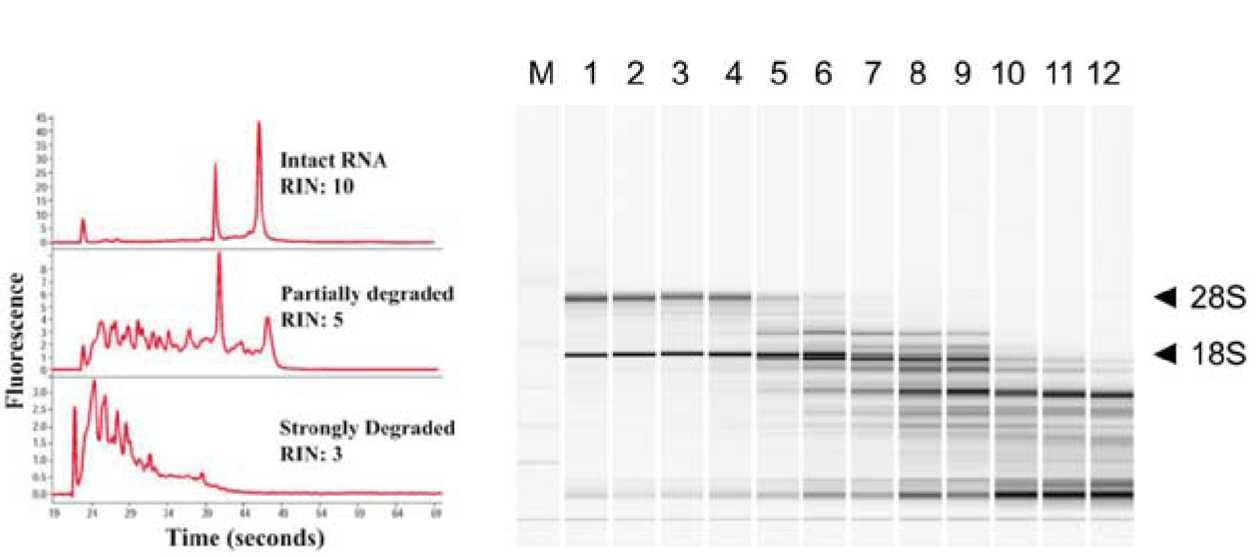
Low RIN
High RIN
- rRNA makes up a large (~85%) of our RNA
- Based on the ratio of 28S and 18S rRNA vs. all RNA
After today, you should have a better understanding of
The role of RNA-seq in modern transcriptomics
Microarrays
Once upon a time, we had microarrays
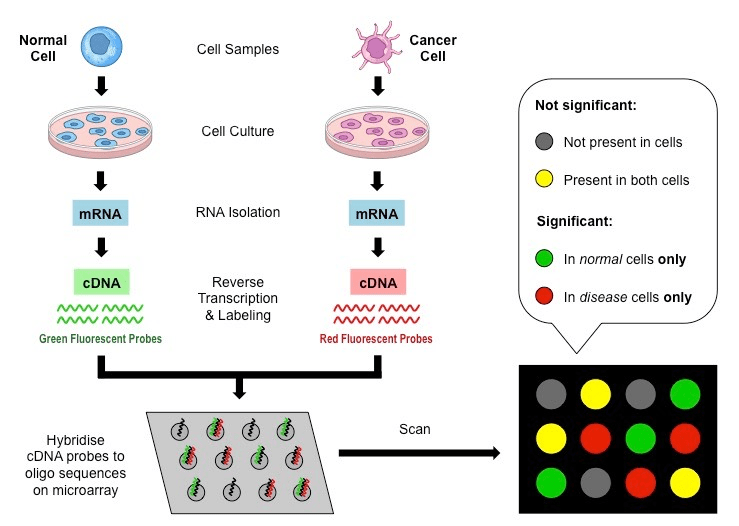
(Now obsolete)
Microarrays have some caveats
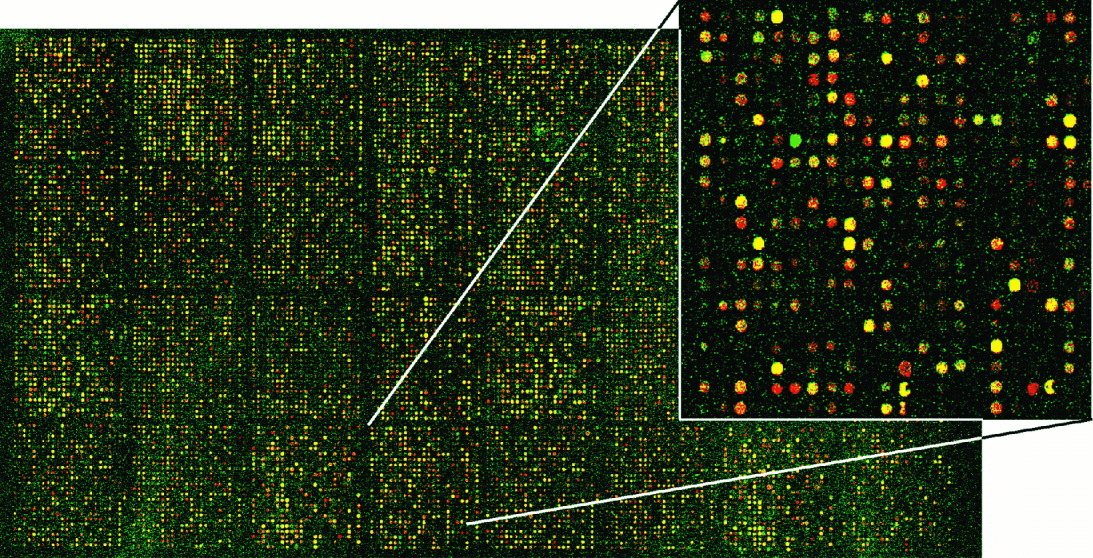
- Limited to known sequences: Can only detect pre-defined sequences
- Cross-hybridization: Similar sequences may cause false positives
- Limited dynamic range: May miss very low or high abundance transcripts
- Normalization challenges: Complex process, potential for bias

After today, you should have a better understanding of
The role of RNA-seq in modern transcriptomics
RNA-seq
RNA sequencing changed the game
Now we just sequence the cDNA
- RNA-seq doesn't require prior knowledge of sequences
- Enables discovery of novel transcripts and isoforms
- Provides absolute quantification rather than relative concentration
Advantages

TopHat questions
What is the primary advantage of RNA-seq over microarray technology?

Which sample has a higher RIN?
After today, you should have a better understanding of
Why read mapping is essential for transcriptomics
Read Mapping is Essential for Making Sense of RNA-seq Data
- RNA-seq produces millions of short sequencing reads (~50–150 bp).
- These reads must be correctly mapped to a reference genome or transcriptome to identify which genes are being expressed.
- Proper alignment enables:
- Gene expression quantification – Counting mapped reads per gene.
- Isoform detection – Distinguishing different transcript variants.
- Splicing analysis – Identifying exon-exon junctions.
The Goal of Read Alignment is to Reconstruct Gene Expression Patterns
- Each RNA-seq read represents a small fragment of a transcript.
- By mapping reads to a reference genome or transcriptome, we can:
- Identify which genes are active in a sample.
- Measure the relative abundance of different transcripts.
- Detect novel isoforms and alternative splicing events.
- Without accurate alignment, downstream analysis (e.g., differential expression) is unreliable.
Challenges in Aligning Short Reads to a Large Reference Genome
The human genome is ~3 billion bases, but RNA-seq reads are only ~100 bases long.
A naïve approach would require searching for every read across billions of bases, which is computationally infeasible.
Why is this a problem?
- Reads may match multiple locations (repetitive sequences).
- Sequencing errors create mismatches, making alignment difficult.
- Polymorphisms & mutations in different individuals affect alignment accuracy.
Why Transcriptomic Read Mapping is Different from Genomic Read Mapping
- Unlike DNA sequencing, RNA sequencing includes spliced transcripts.
-
Key problem: Reads from mRNA span exon-exon junctions, but the genome contains introns.
- Example: A read from an mRNA transcript might originate from exon 1 and exon 2, but the genome has a large intron in between.
- Consequence: Traditional genomic aligners fail to align these reads correctly.
- Solution: Transcriptomic aligners must allow for gapped alignments that bridge exon-exon junctions.
Splice Junctions Complicate Read Mapping
- Genes contain introns that are removed during splicing, but these are not present in mature RNA.
- Example: A 100-bp RNA-seq read may contain 50 bp from Exon 1 and 50 bp from Exon 2, skipping a 10,000-bp intron.
-
Challenges:
- Reads spanning junctions do not exist as contiguous sequences in the genome.
- Aligners must infer intron-exon boundaries from known or novel splice sites.
- Specialized RNA-seq aligners (e.g., STAR, HISAT2) are designed to handle this.
Isoforms Add Another Layer of Complexity to Read Mapping
- A single gene can produce multiple transcript isoforms through alternative splicing.
- This means a single RNA-seq read might belong to:
- One specific isoform.
- Multiple overlapping isoforms.
- An unannotated, novel isoform.
- Consequence: Read mapping must account for multiple possible alignments within the same gene.
Before the next class, you should
Lecture 06B:
Read mapping -
Methodology
Lecture 06A:
Read mapping -
Foundations
Today
Thursday
BIOSC 1540: L06A (Read mapping)
By aalexmmaldonado
BIOSC 1540: L06A (Read mapping)
- 237



