iTag sequencing
Adam R. Rivers
Microbial genomics and metagenomics workshop
April 2016

DW Waite, P Deines, MW Taylor. 2012. Gut Microbiome of the Critically Endangered New Zealand Parrot, the Kakapo (Strigops habroptilus). PLOS One. v. 7, p. e35803.

Gustav Klimt, "Tree of life", 1909, Wikimedia
Amplicon sequencing at JGI
Bacterial and Archaeal 16S rRNA
Eukaryotic 18S rRNA
Fungal Internally Transcribed Spacer (ITS)
5.8S
18S
28S
18S
16S
V4-V5
V4
ITS2

Questions iTags answer most often
- Taxonomic composition
- Sample similarity
- Alpha diversity
- Beta diversity
- Community structure
- Imputed function
iTags sequencing is transitioning from descriptive to experimental applications





Langille et al. 2013
An overview of iTags sequencing at JGI
10,000
samples in 2016
4 Billion
iTags reads per year
- Production scale sequencing
- An automated QC system
- The iTagger automated analysis system
- Archival data storage
- Public web portal
Library creation and sequencing



Two 96 well plates
PCR with 1 forward and 196 reverse primers
-
Illimina MiSeq
-
2 X 300 bp
-
45-50M reads
-
350,000 reads per sample

Multiplexing

Illumina iTags primer layout
5'-
-3'
Forward primer 515Y-F
Illumina adapter
Barcode
Spacer
Primer
AATGATACGGCGACCACCGAGATCTACAC TCTTTCCCTACA CGACGCTCTTCCGATCT NNNNN GTGYCAGCMGCCGCGGTAA
Linker
Illumina adapter
Barcode
Spacer
Primer
AGCAGAAGACGGCATACGAGAT AGCGAACCTGTT GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT CTA CCGYCAATTYMTTTRAGTTT
Linker
Reverse primer 926-R
Current primers used
Bacterial and Archaeal 16S rRNA
Eukaryotic 18S rRNA
Fungal Internally Transcribed Spacer (ITS)
5.8S
18S
23S
18S
16S
V4-V5
V4
ITS2
515YF GTGYCAGCMGCCGCGGTAA Parada et al. 2015
926R CCGYCAATTYMTTTRAGTTT Parada et al. 2015
565F CCAGCASCYGCGGTAATTCC Stoeck et al. 2010
948R ACTTTCGTTCTTGATYRA Stoeck et al. 2010
ITS9F GAACGCAGCRAAIIGYGA Menkis et al. 2012
ITS4R TCCTCCGCTTATTGATATGC White et al. 1990
Updates to iTags sequencing
- V4-V5 region 515YF - 926R (Parada et al. 2015)
- Change to new annotation databases:
- Major version release of iTagger 2.0


Locked primers for symbiosis
In Host dominated communities their DNA predominate

iTagger: Read quality control
Read QC
OTU Clustering
Taxonomic analysis
- Remove contaminants (JGI RQC)
- Merge reads (Usearch)
- Primer trim (Usearch)
- Size and expected error filtering
- Dereplication
| Total Reads | 40 M |
|---|---|
| PhiX | 0.3% |
| Illumina artifacts | 0.1% |
| Primer trimmed | 5% |
| Mate pair extended | 2% |
| Length Filtered | <0.1% |
| Final quality filtering | 15% |
| Final Reads | 26M |
| Average seqs per sample | 364k |
iTagger: OTU clustering
Read QC
OTU Clustering
Taxonomic analysis
- Reads are sorted by abundance
- USEARCH-Uparse is used to cluster iteratively at 99-97%


Edgar (2013)
iTagger: Taxonomic analysis
Read QC
OTU Clustering
Taxonomic analysis
Utax classifier classifies centroids using 8-mers and a principled training model

Precision
Error precision curve for V3-V5
Summary plots (QIIME)
Making connections across data
JGI has one of the largest 16S repositories. We have created a database to record and compare V4-V5 OTUs across projects.


We are investigating adding visualization tools to explore iTag data.
Bik et al. 2014 (preprint)
Genome portal demonstration
Going further with iTags data
- General tools: Qiime, Mothur
- 16S pipeline: JGI's iTagger, SilvaNGS
- OTU clustering: Qiime, Mothur, UPARSE
- Functional imputation: Picrust, Tax4Fun
- Databases for 16S data: RDP, Greengenes, Silva
- Repositories for 16S data: Qiita
- Statistical testing: Vegan, LEfSe, Phyloseq,
- Network analysis: Cytoscape, Statnet, Spiec-easi, Bnlearn
Future directions: Full length reads
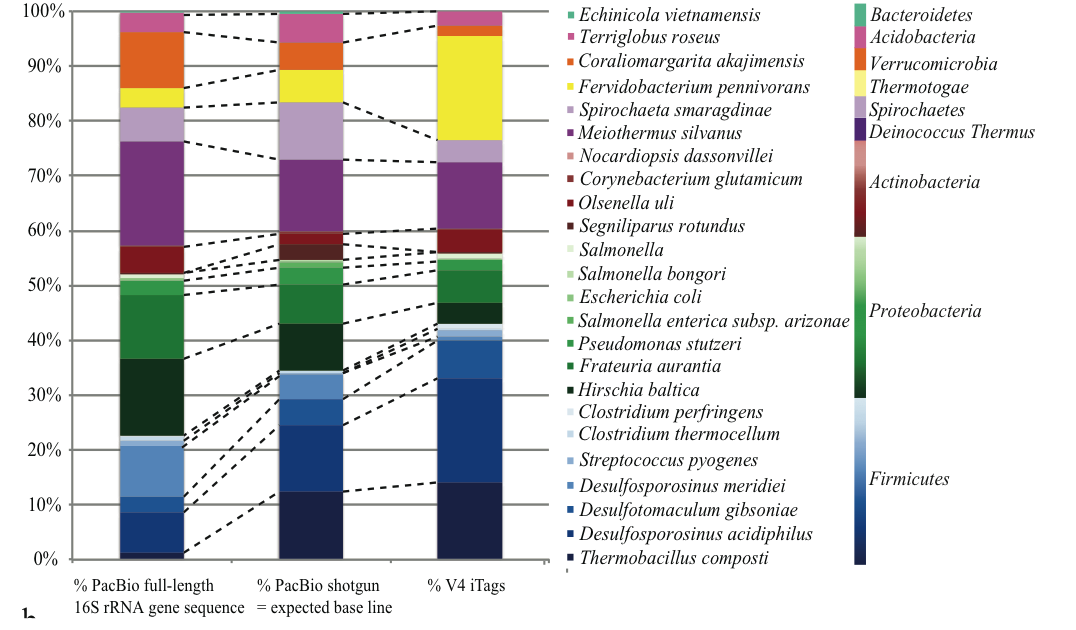
Future directions: Ecological networks

Future directions: Ecological networks

A bayesian network constructed from 350 wetland samples
Directed links indicate conditional dependence
Tringe and Theroux, unpublished
estimated with BNlearn
Questions?
Adam R. Rivers
arrivers@lbl.gov
iTags Presentation
By Adam Rivers
iTags Presentation
- 1,280

