Genome structure
from Hi-C
Sasha Galitsyna
Skoltech
2019
Hi-C review
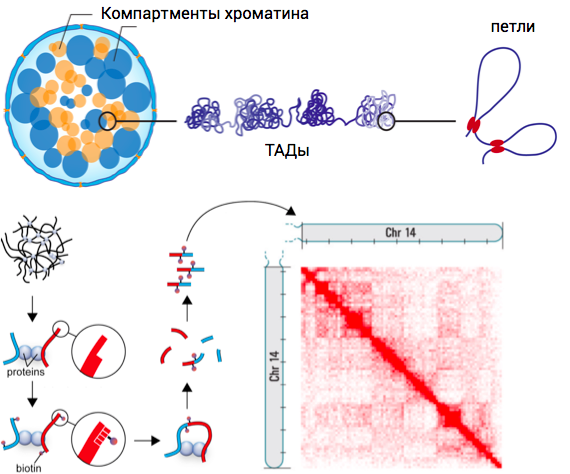
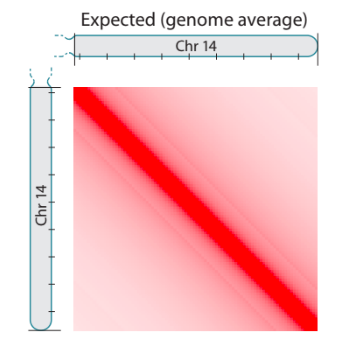
Hi-C: factors contributing
Physical:
- Scaling
- Rabl-configuration (if present)
- Compartments
- TADs
- Loops
Technical:
- Ligation radius, restriction effectivity
- DNA accessibility
- PCR amplification
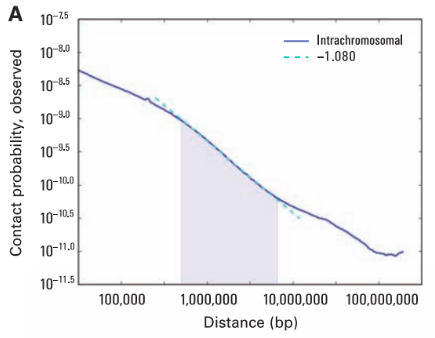
Scaling
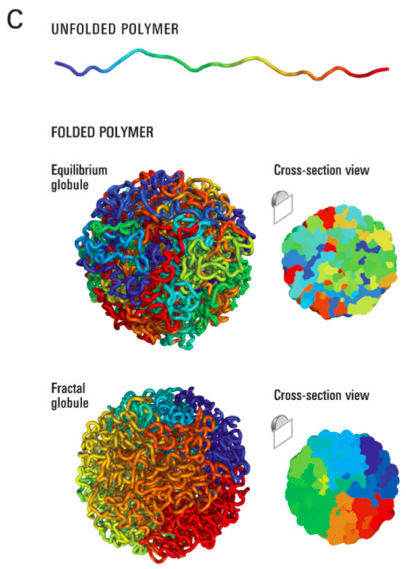
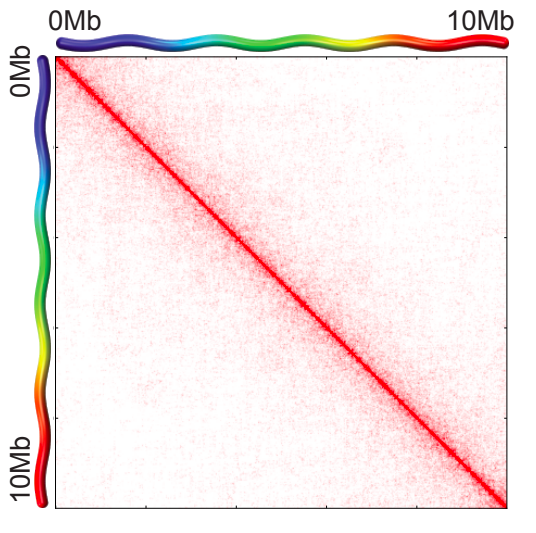
Scaling
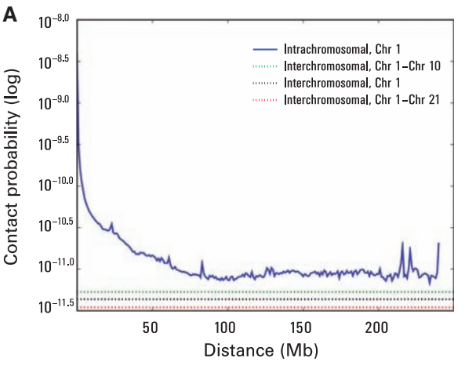
Rabl configuration
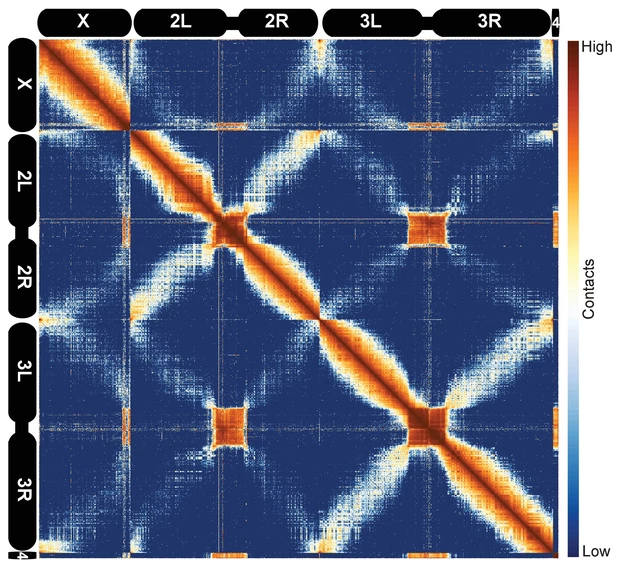
Stadler eLIFE 2017
Other high-order structures
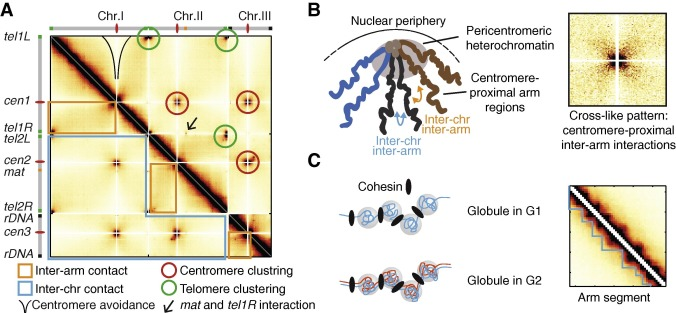
Fission yeast:
Other high-order structures
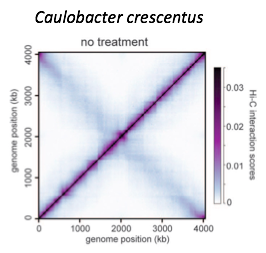
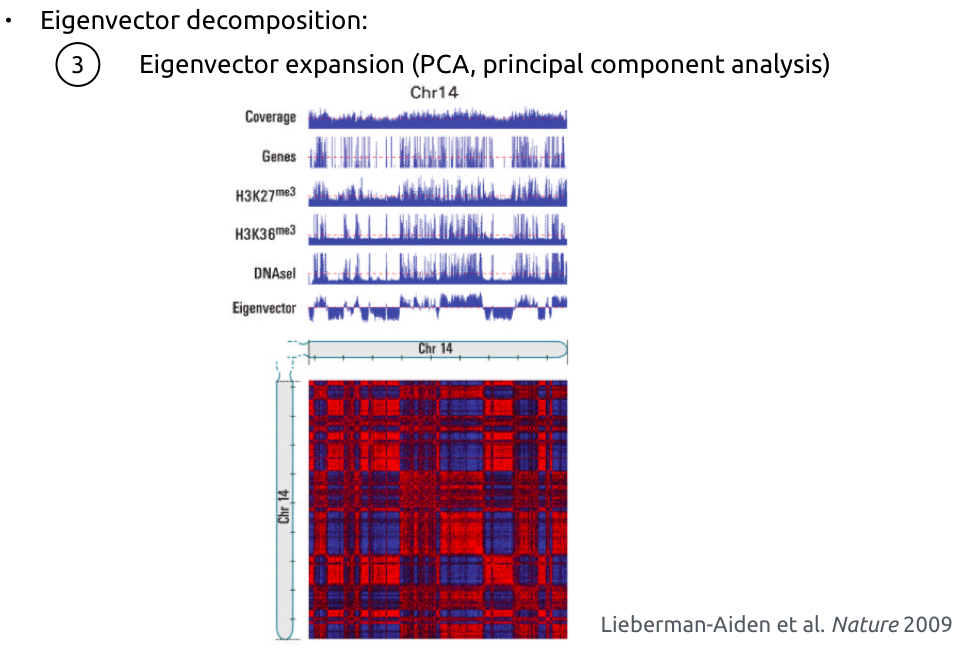
Compartments calling
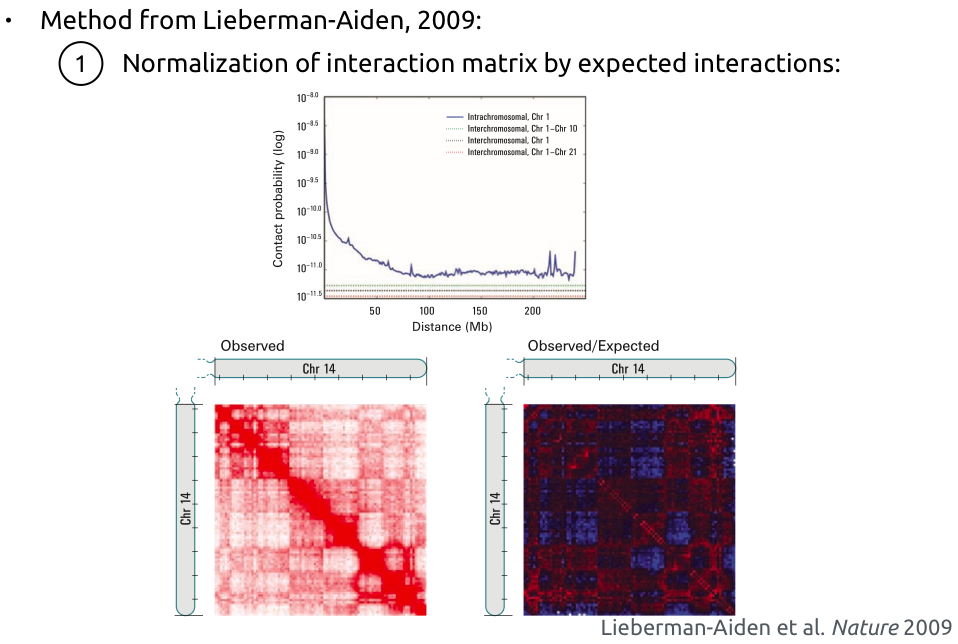
Compartments calling
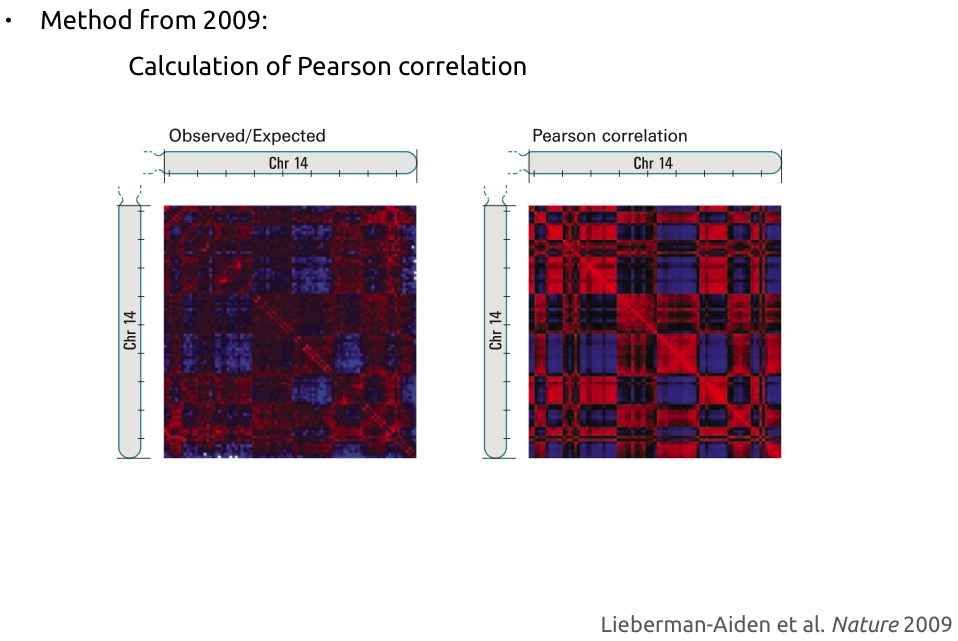
Compartments calling
TADs calling
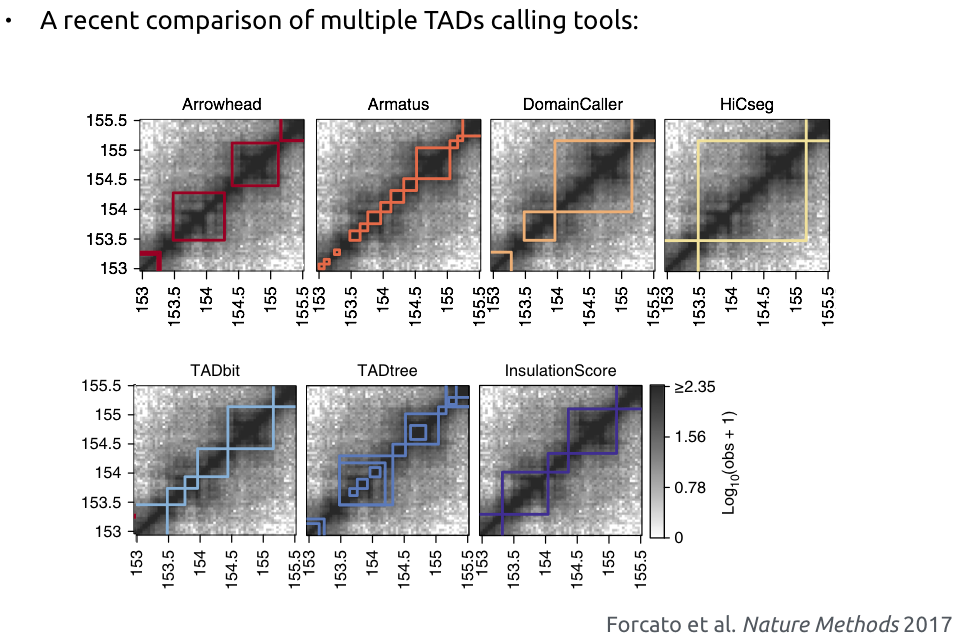
TADs calling
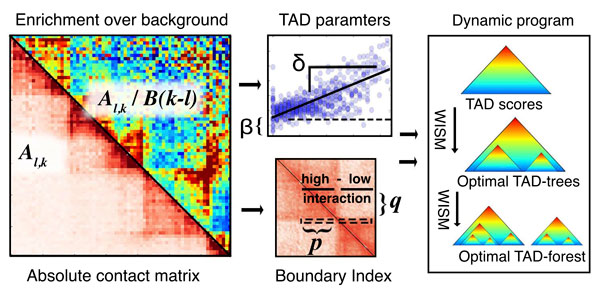
TADTree example:
Loops
Invariant Hi-C interactions patterns
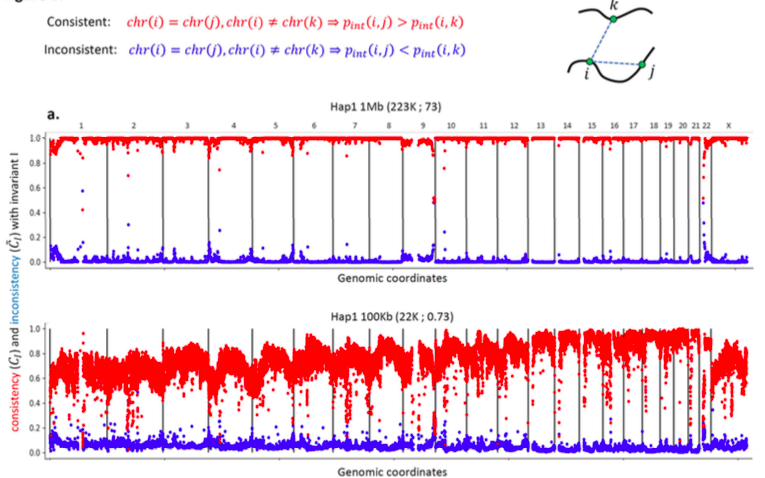
Invariant pattern I: Intrachromosomal interaction enrichment
Oddes 2018
Invariant Hi-C interactions patterns
Invariant pattern II: Distance-dependent interaction decay
Oddes 2018
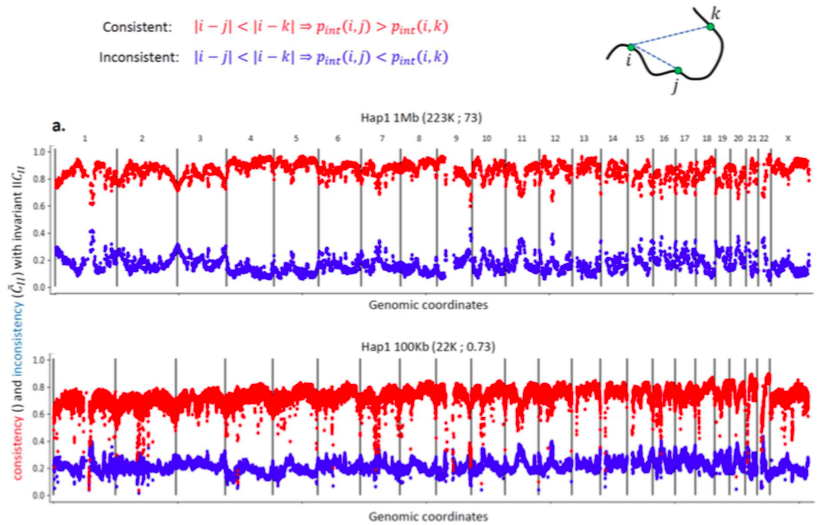
Invariant Hi-C interactions patterns
Invariant pattern III: Local interaction smoothness
Oddes 2018
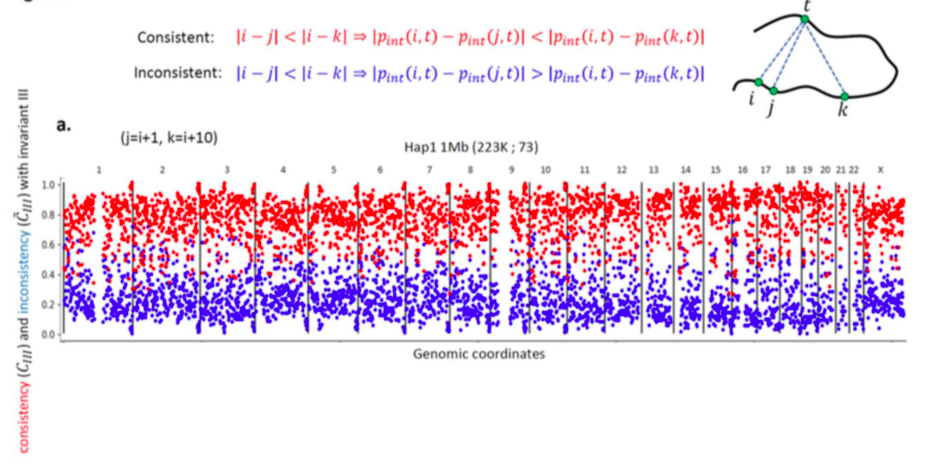
Invariant Hi-C interactions patterns
Oddes 2018
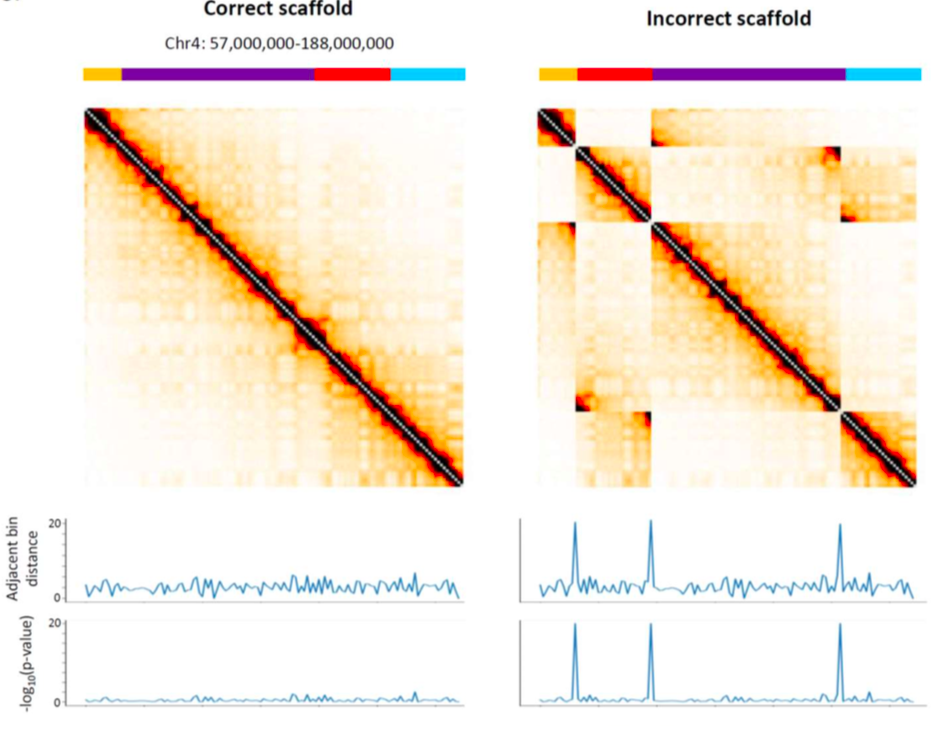
Invariant Hi-C interactions patterns:
low resolution
Oddes 2018
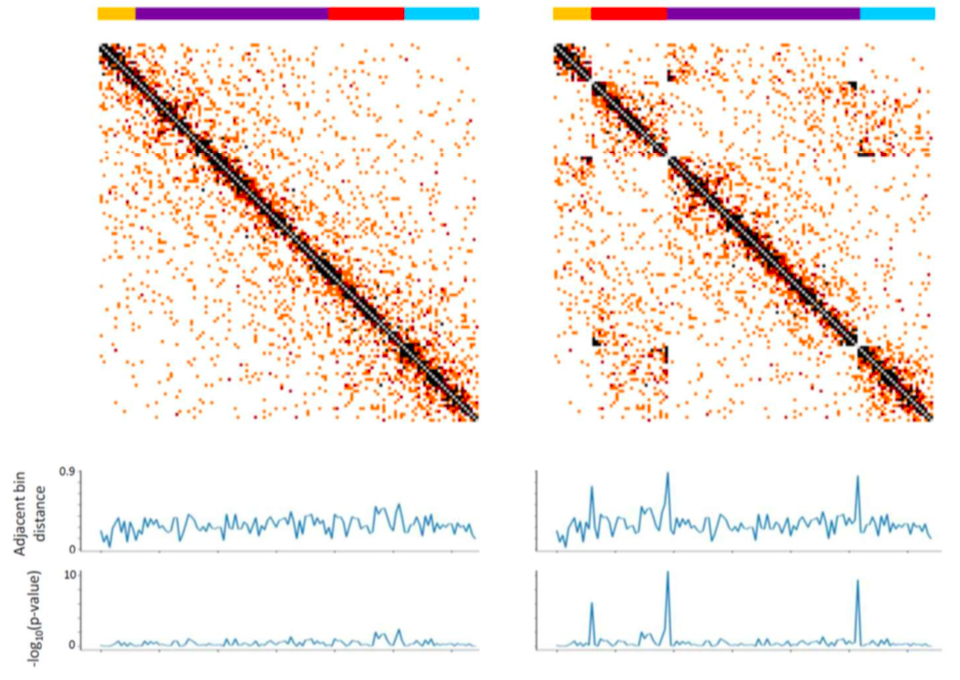
genomes scaffolding
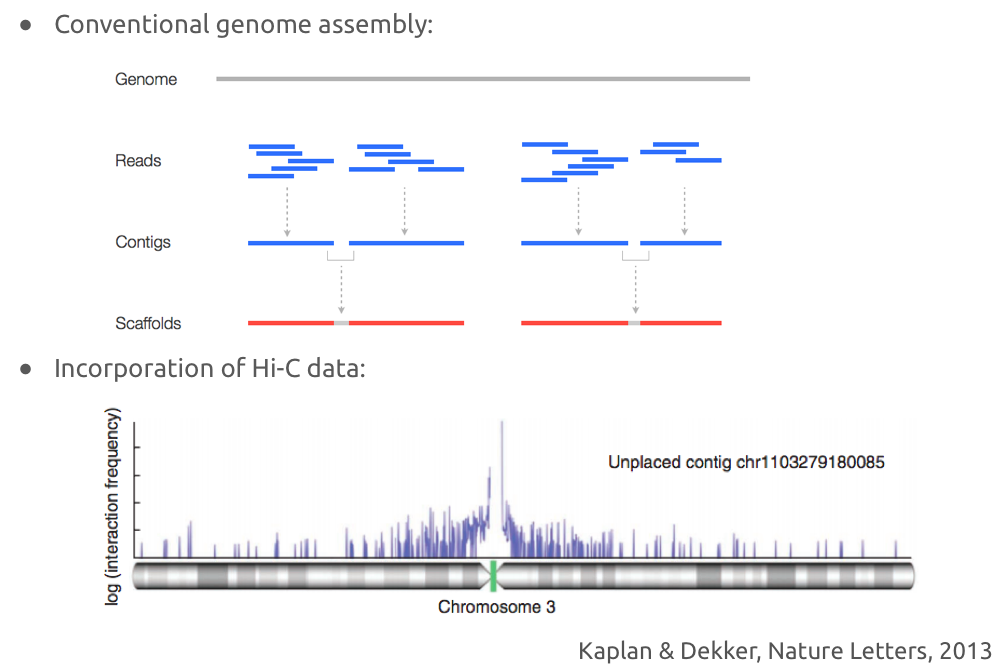
genomes scaffolding with Hi-C

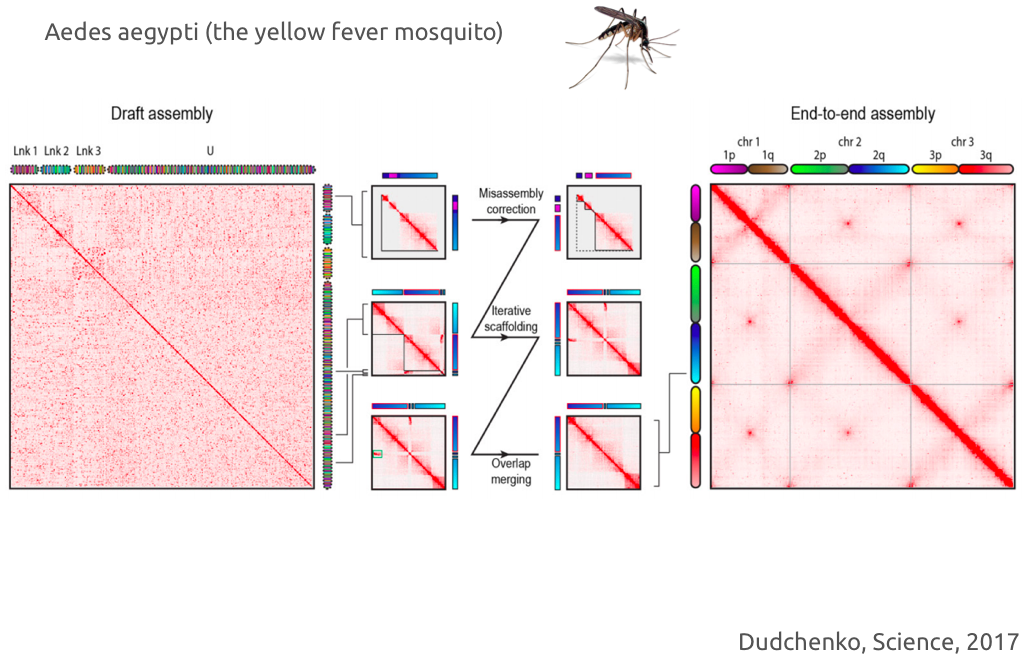
genome assembly: examples
References
-
"Three invariant Hi-C interaction patterns: applications to genome assembly"
Oddess et al. 2018 bioRxiv 10.1101/306076 - Kaplan lab -
"The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000"
Dudchenko et al. 2018 bioRxiv 10.1101/254797 - Aiden lab -
"De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds"
Dudchenko et al. 2017 Science 10.1126/science.aal3327 - Aiden lab -
"Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours"
Harewood et al. 2017 Genome Biology 10.1186/s13059-017-1253-8 - Fraser lab -
"Chromatin conformation analysis of primary patient tissue using a low input Hi-C method"
Diaz et al. 2018 NatCom 10.1038/s41467-018-06961-0 - Vaquerizas lab -
"Improved reference genome of Aedes aegypti informs arbovirus vector control"
Matthews et al. 2018 Nature 10.1038/s41586-018-0692-z -
"Genome organization and DNA accessibility control antigenic variation in trypanosomes"
Muller et al. 2018 Nature 10.1038/s41586-018-0619-8
hic_assembly2019
By agalicina
hic_assembly2019
- 426



