OMICS DATA ANALYSIS
Midterm evaluation

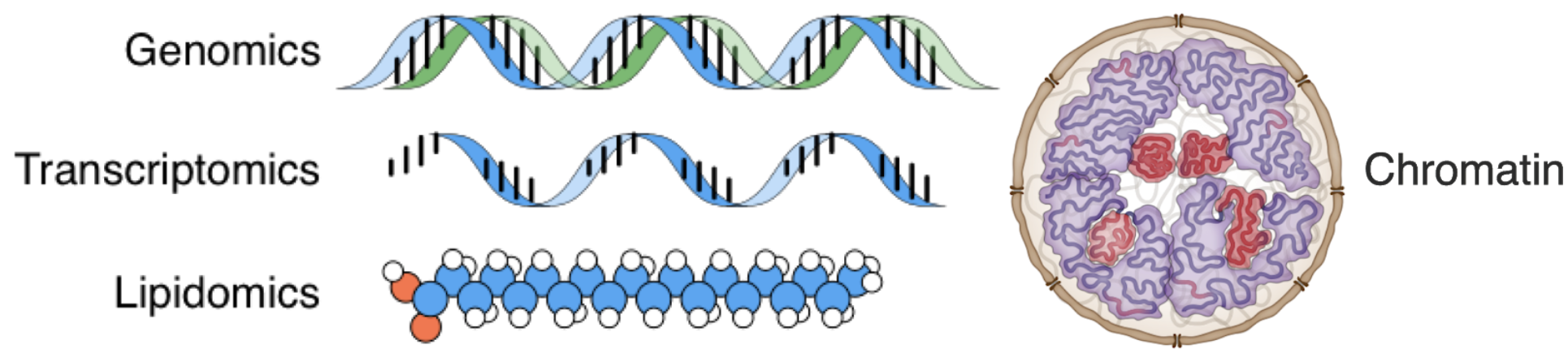
How we get tired...






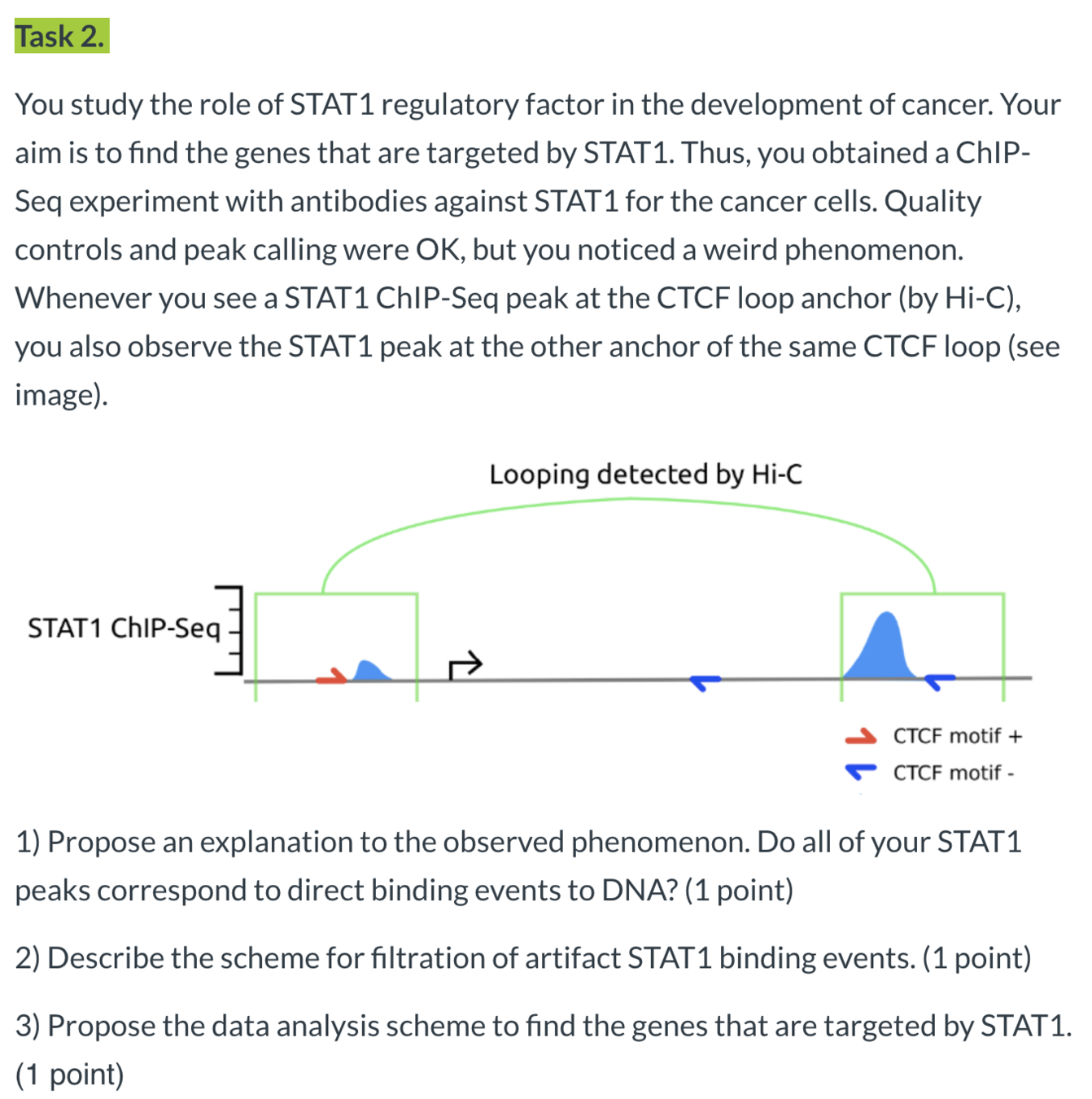
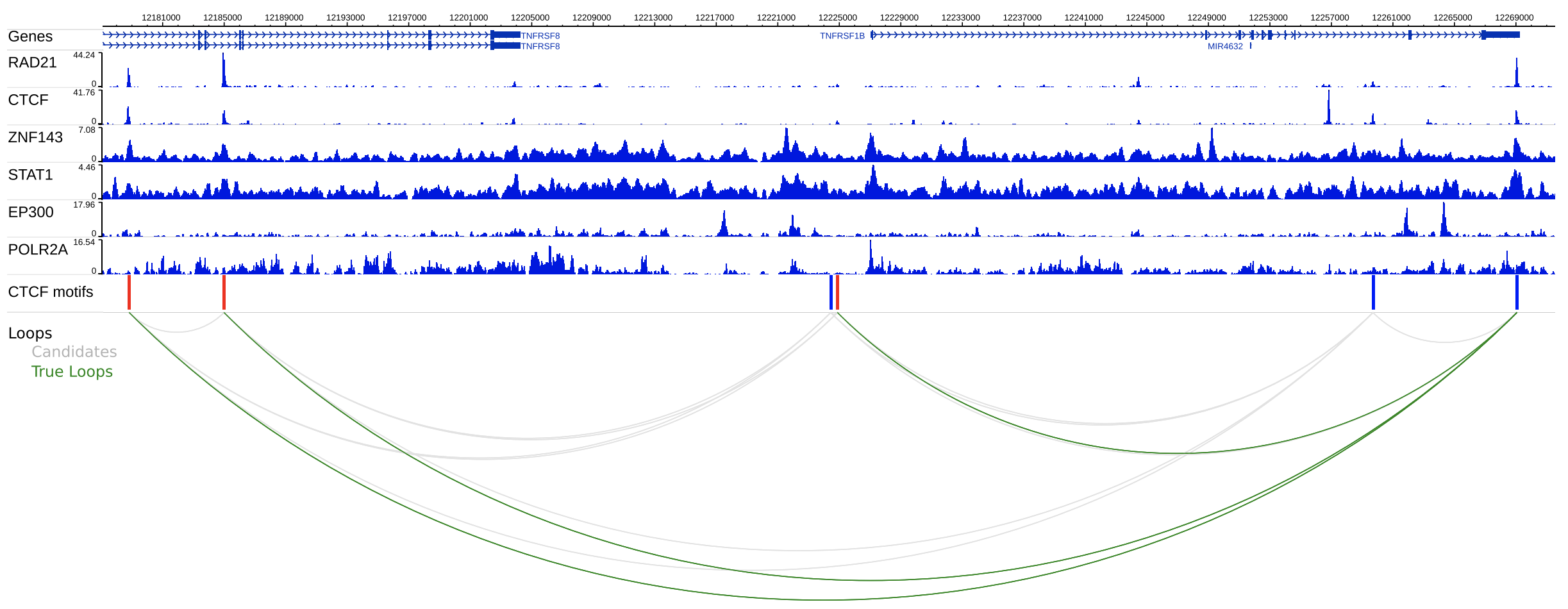
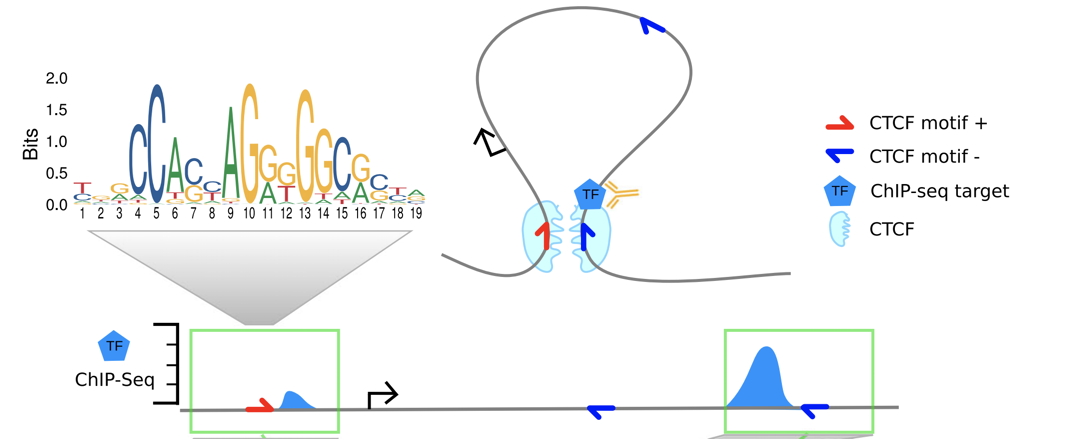
Whenever you see a "STAT1" ChIP-Seq peak at the CTCF loop anchor (by Hi-C), you also observe the STAT1 peak at the other anchor of the same CTCF loop:
Hi-C loops
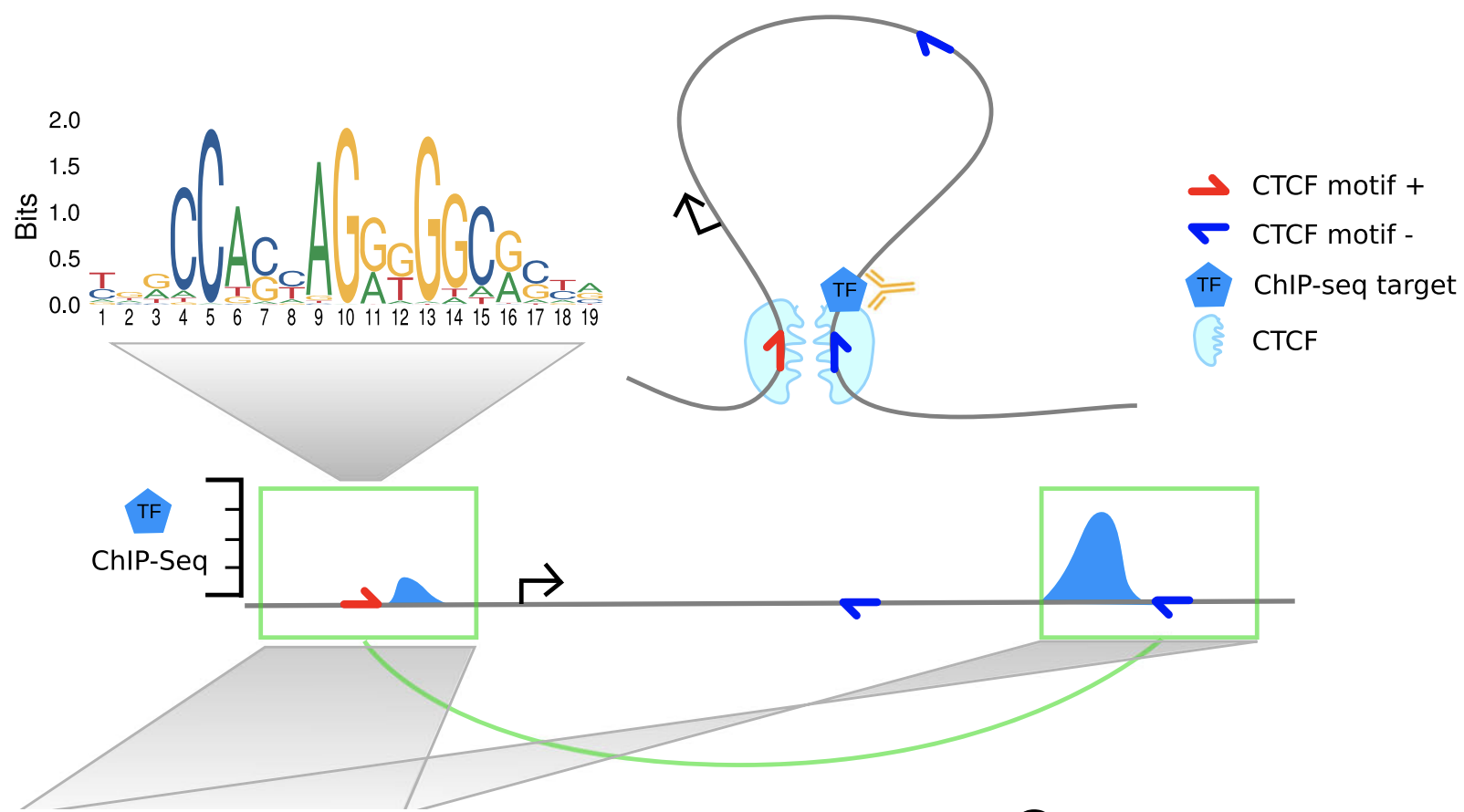
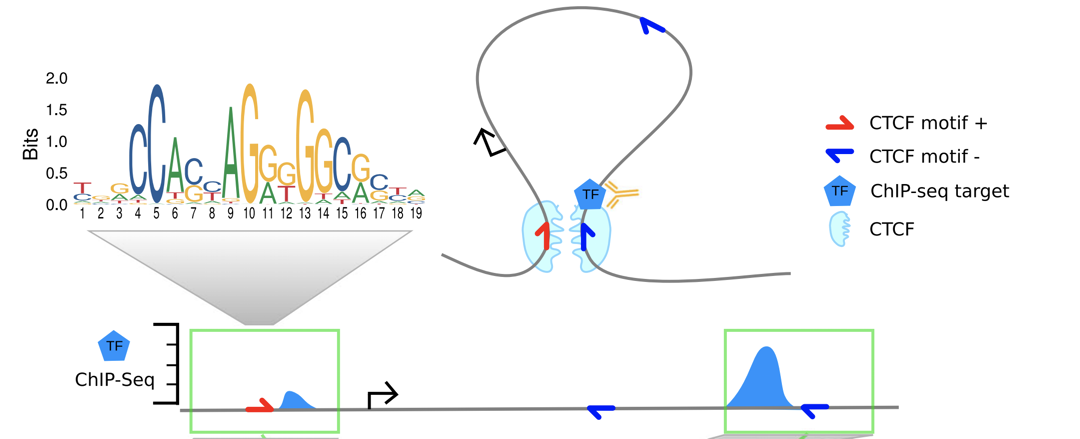
The explanation proposed by the authors:
"While ChIP-seq was not designed to detect chromatin interactions, the formaldehyde treatment in the ChIP-seq protocol cross-links proteins with each other and with DNA. Consequently, also regions that are not directly bound by the targeted TF but interact with the binding site via chromatin looping are coimmunoprecipitated and sequenced. This produces minor ChIP-seq signals at loop anchor regions close to the directly bound site."
Alternative explanations proposed by you:
- Dimerization of "STAT1",
- CTCF-"STAT1" protein complex,
- "STAT1" complex with some other factors that co-localize with loops,
- Genome duplications,
- Mutations in cancer,
- Problems with sample preparation, e.g. binding of antibodies to all open chromatin regions.
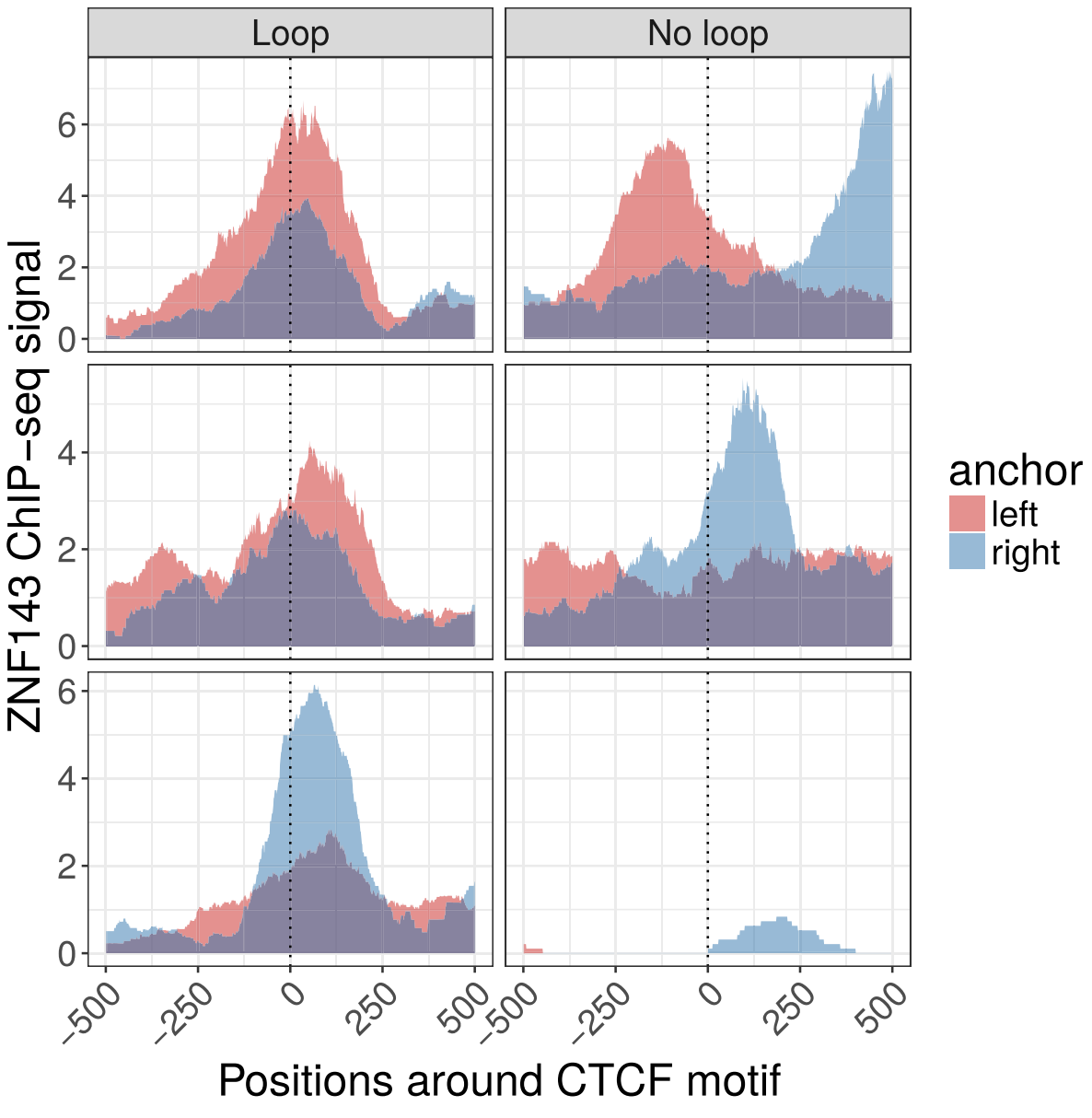
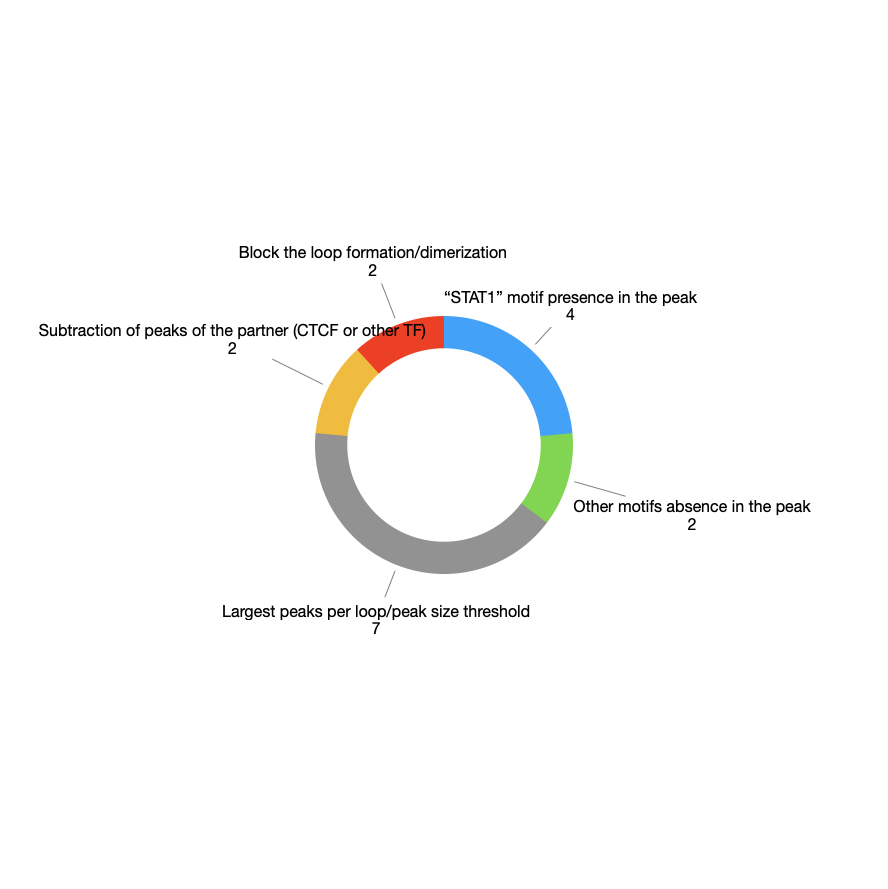
Filtration of direct binding events strategies:
ATAC-Seq
Hsu et al. 2018
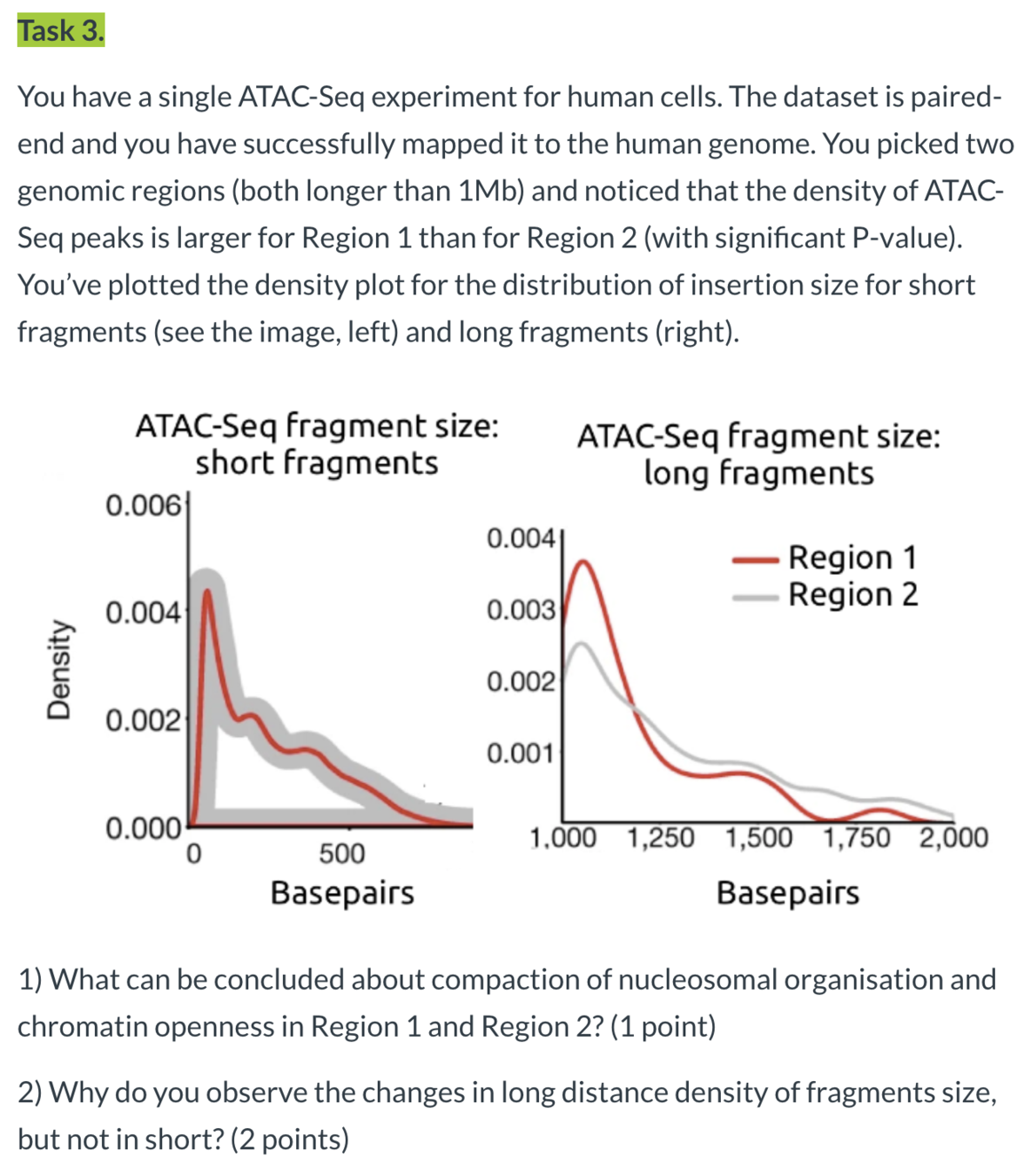
paired-end sequencing and mapping
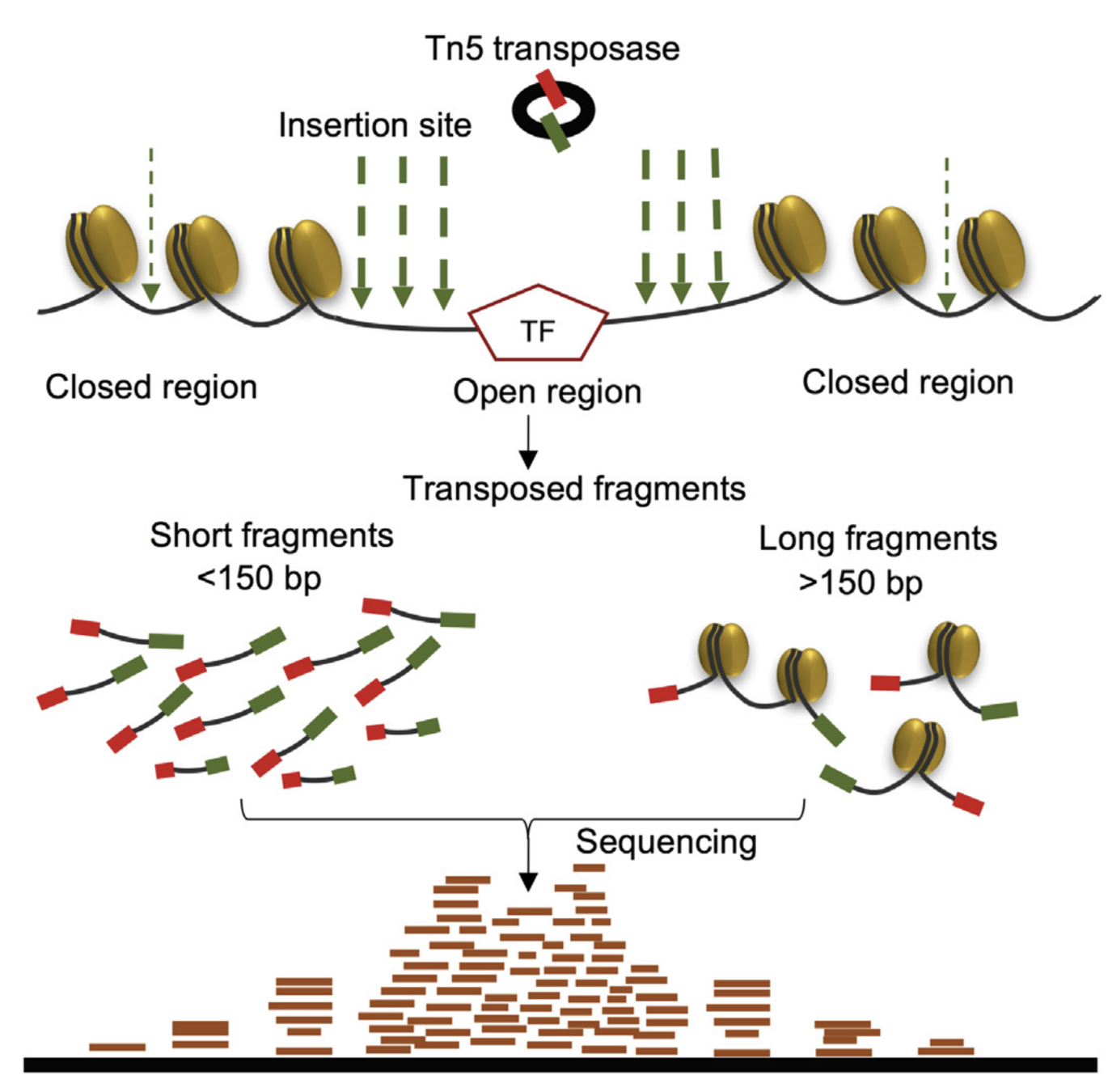
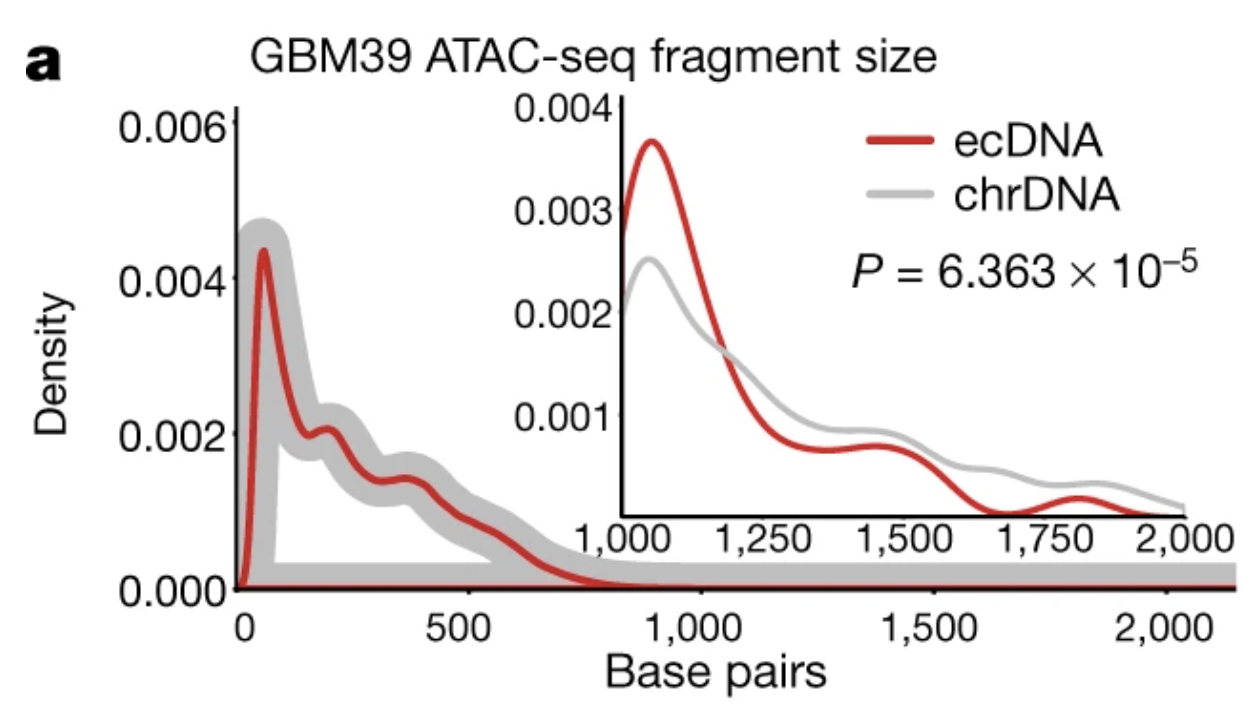
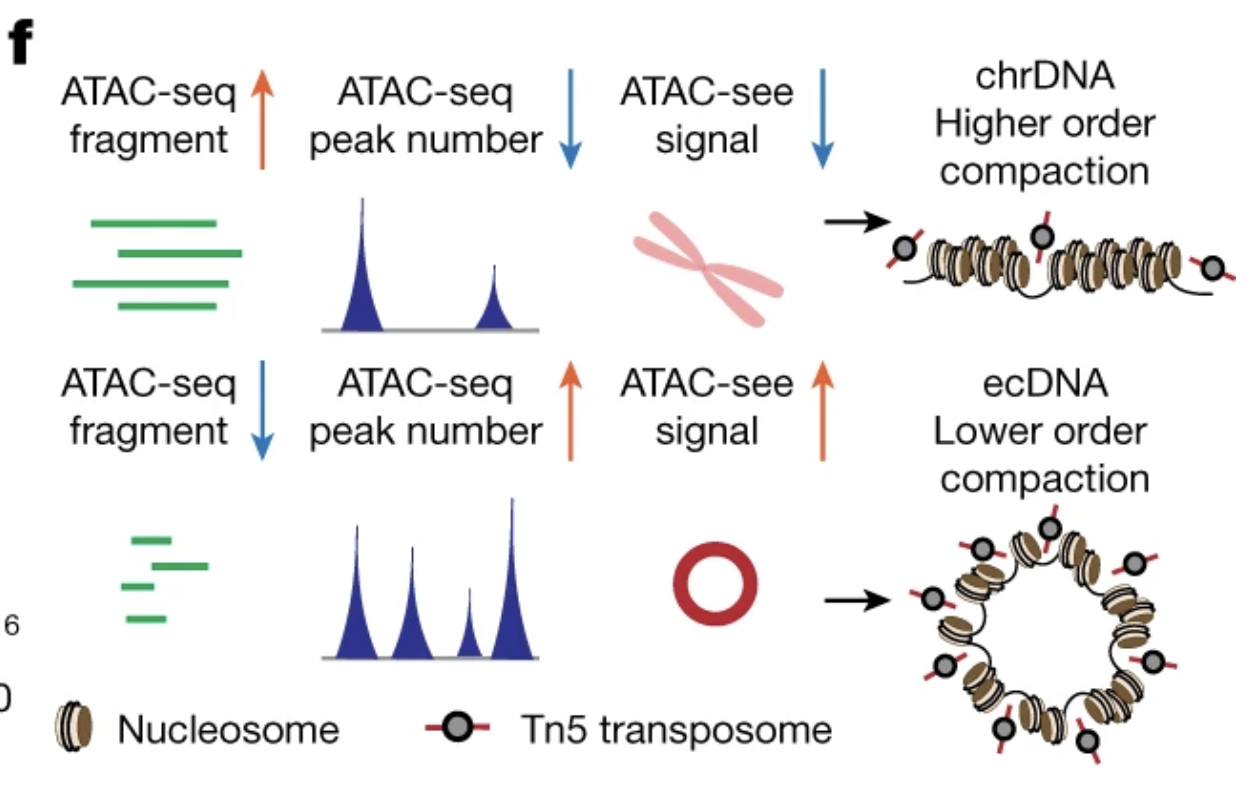
"ecDNA displayed a significant deficit in the number of long fragments (more than 1,200 bp) from ATAC-seq and MNase-seq, indicative of compacted nucleosomal arrays, and a significantly increased number of ATAC-seq peaks, which suggests that the ecDNA chromatin landscape is more accessible than chrDNA, because its nucleosomal organization is less compacted."
Red-C for RNA-DNA high-throughput assay
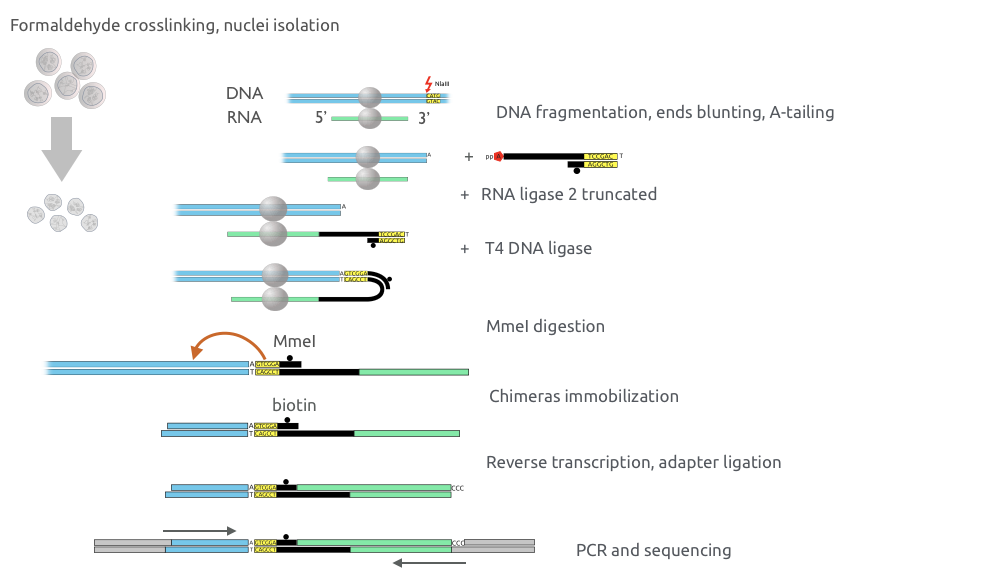
Gavrilov, Zharikova, Mironov, Galitsyna, paper in review
Red-C for RNA-DNA high-throughput assay
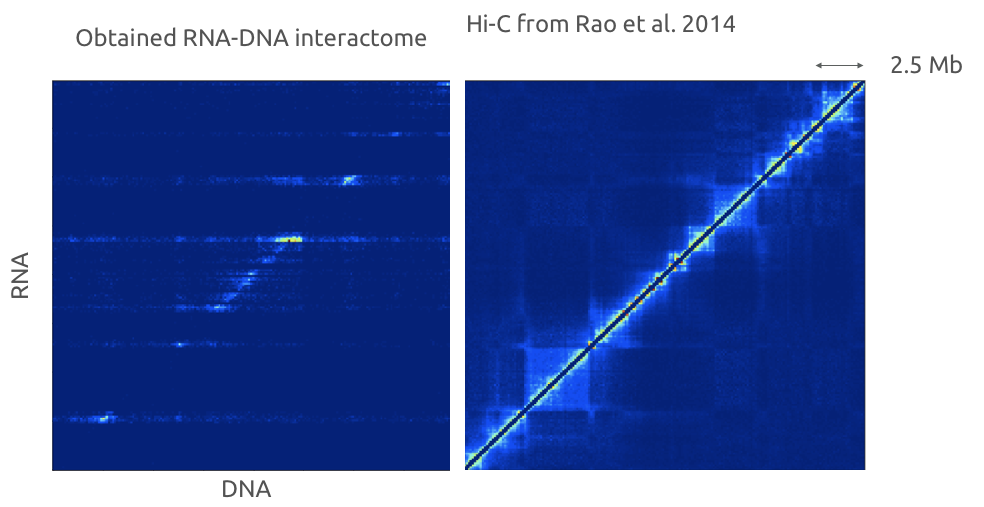
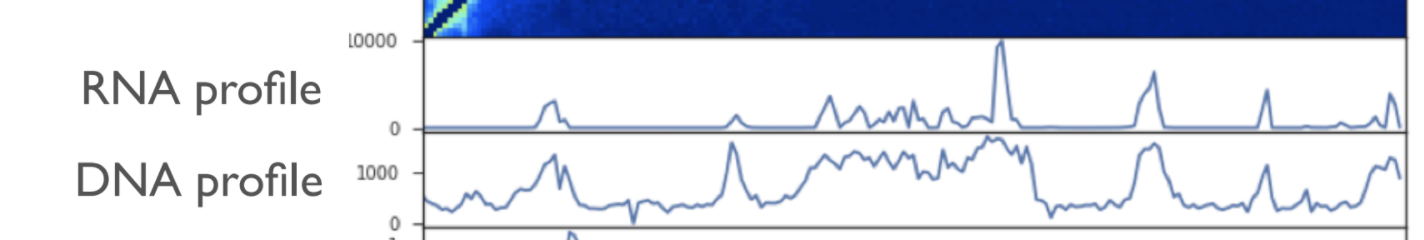
Typical mistake is that you don't consider that coverages for RNA and DNA are separate:
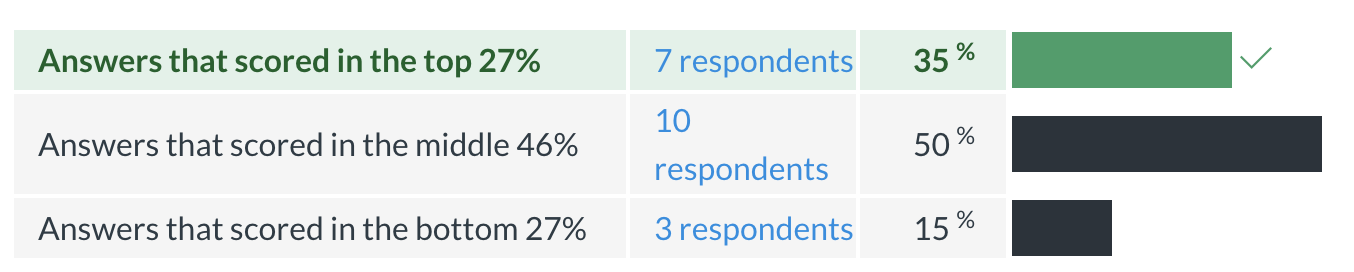
Midterm evaluation results
By agalicina
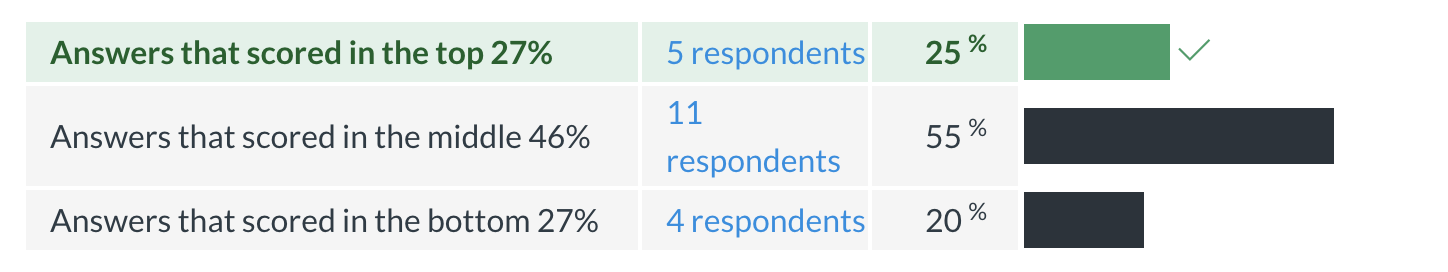
Midterm evaluation results
Results of the Omics Data Analysis course Midterm evaluation.
- 165



