NF-Core: Community-based best practice pipeline development in Nextflow
Alexander Peltzer
Quantitative Biology Center (QBiC) Tübingen


http://bit.ly/ismb2018-nfcore

@alex_peltzer


Outlook
- Challenges in computational biology
- Basic introduction to Nextflow
- Introduction to NF-core project


Challenges: Big Data
- Data in computational biology is
- big (PB scale)
- diverse (sequencing, proteomics, metabolomics ...)
- erroneous (e.g. contains sequencing errors)
We need methods and tools to analyze such data!


Challenges: Software dependencies
Workflows / Pipelines consist of
- various different tools
- typically dozens of individual methods
Complex dependency management!


Challenges: Reproducibility
- Large-scale projects more common today
- 1,000 Genomes Project
- 100,000 Genomes Project UK
- Reproduce results with older data / integrate with newer data
Many paper results are not reproducible!


Nextflow
- Custom DSL (domain-specific language) for
- fast prototyping
- enabling task composition
- easy parallelization
- Self-contained: Containerize tasks (e.g. with Docker)
- Isolation of dependencies: Keep container - rerun analysis at any point!


- Community effort to collect production ready analysis pipelines
- Save time in development, more testing, more updates



Phil Ewels
Alex Peltzer
Sven Fillinger
Andreas Wilm
Maxime Garcia
+ many others!
Tiffany Delhomme









All pipelines adhere to requirements
- Nextflow based
- MIT license
- Software bundled in Docker / Singularity
- Continuous integration testing (e.g. Travis CI)
- Stable release tags
- Common pipeline usage and structure


Optional requirements
- Software bundled in Bioconda
- Optimized output formats (e.g. CRAM)
- Explicit support for cloud environments (AWS)
- Benchmarks for running on such environments


Need help?
- Cookiecutter: To get a skeleton for new pipelines
- Linting app: To check what conforms with nf-co.re
- Gitter: To communicate with the community!


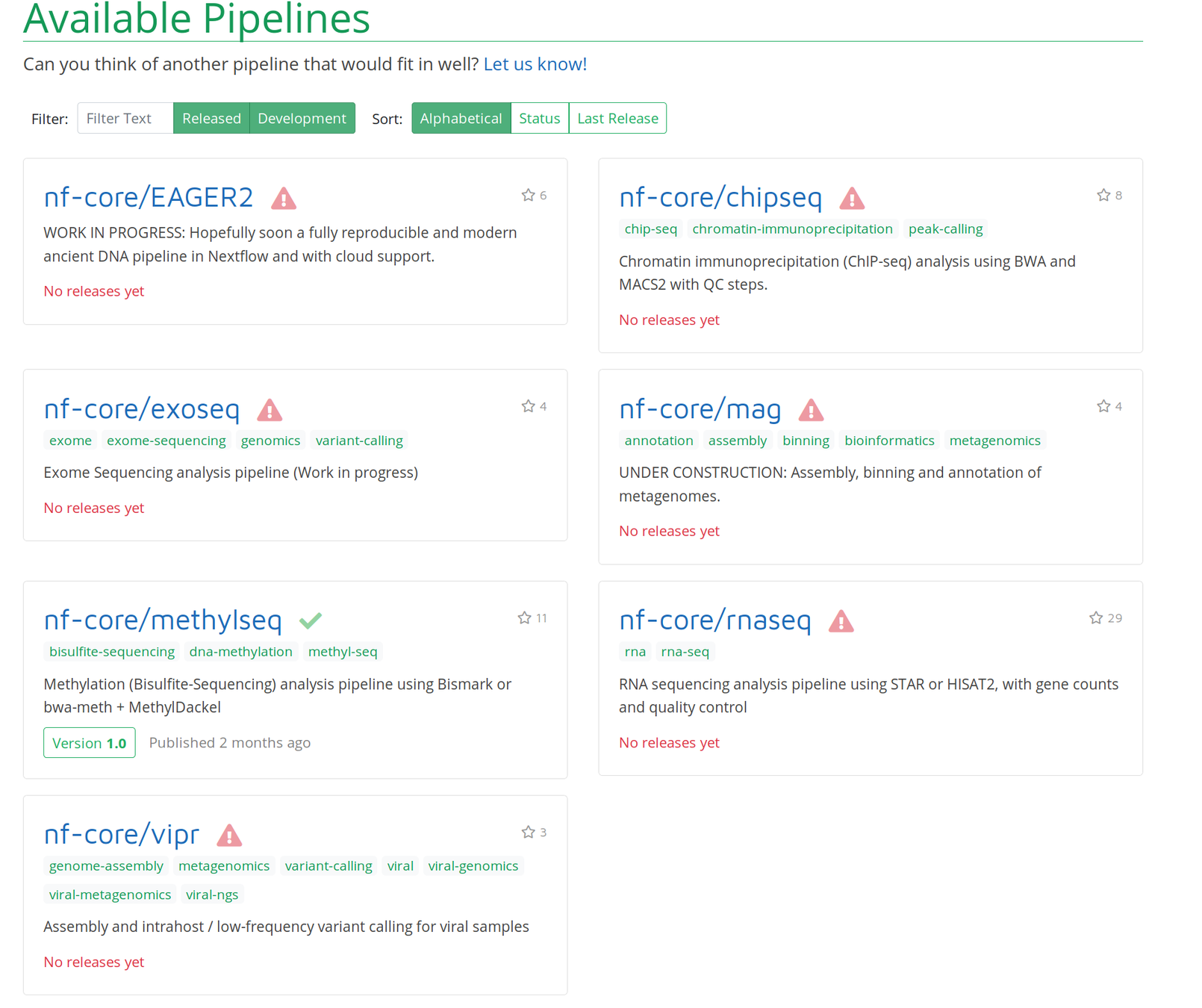


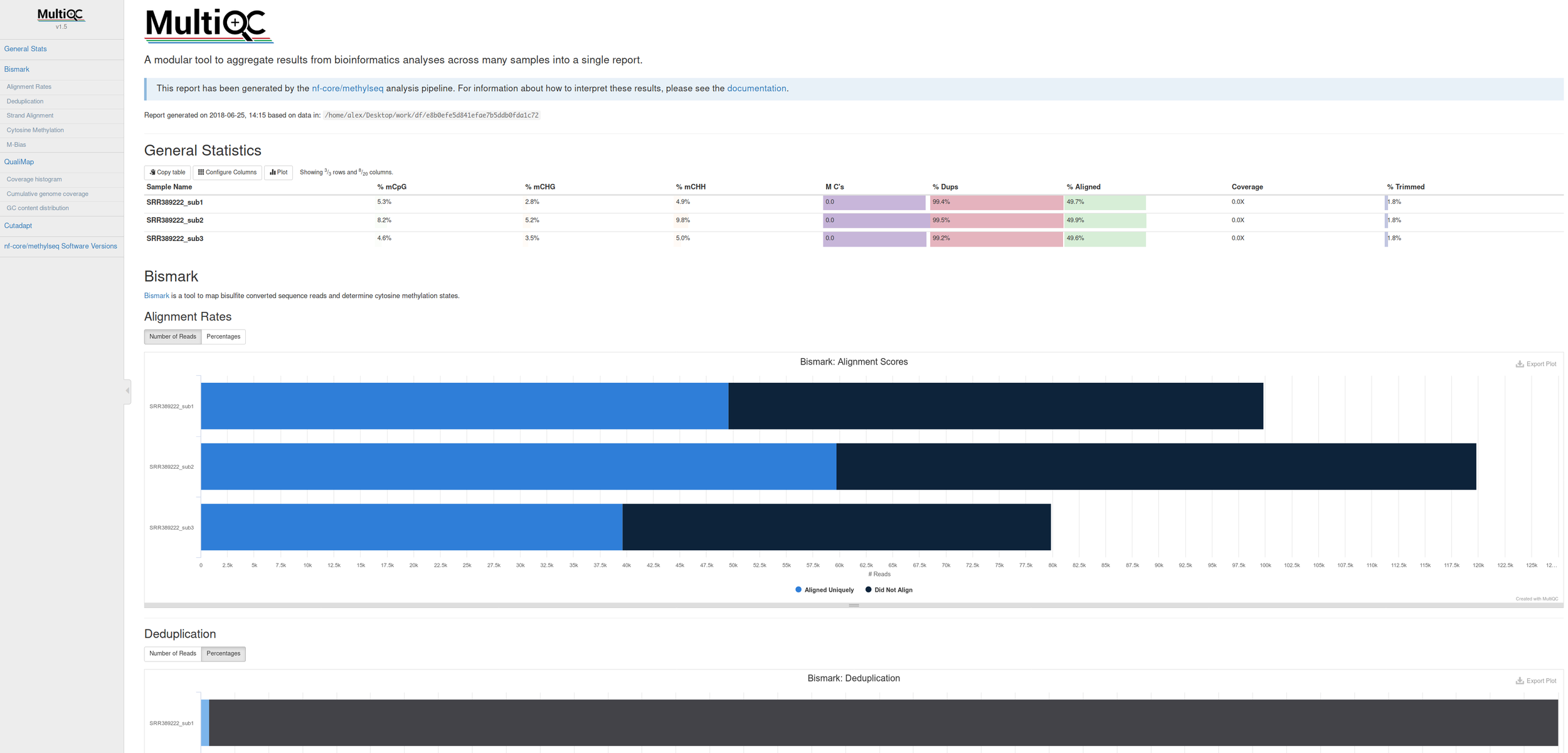
Comes with interactive reports!


Comes with proper documentation!
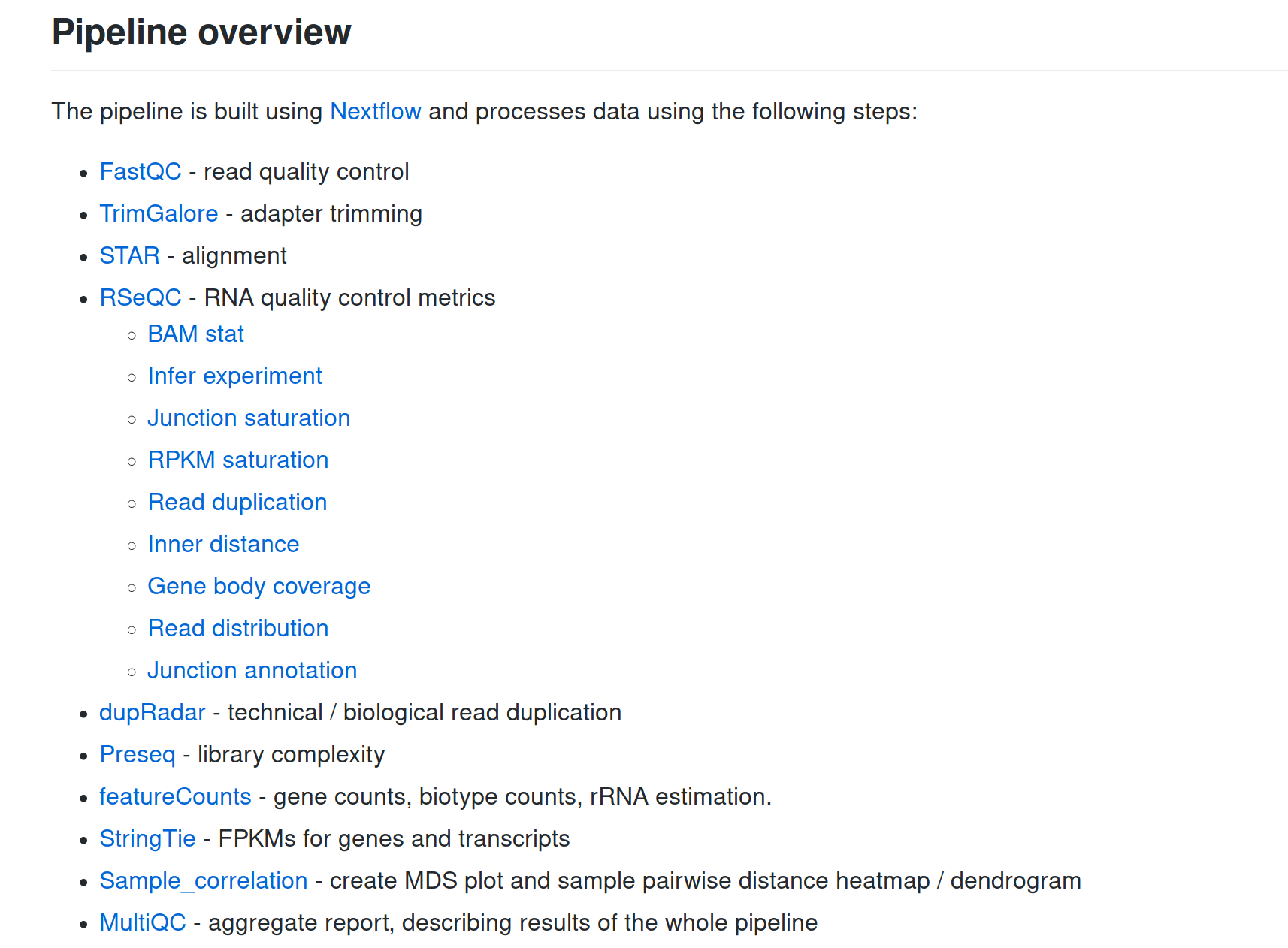


... and a lot more!



Acknowledgements
Phil Ewels (SciLifeLab, Stockholm)
Maxime Garcia (SciLifeLab, Stockholm)
Sven Fillinger (QBiC, Tübingen)
Paolo di Tommaso (CRG, Barcelona)
Evan Floden (CRG, Barcelona)
Andreas Wilm (A* Singapore, Singapore)
Tiffany Delhomme (IARC, Paris)
ISMB Bioinfo Core Workshop NF-Core
By Alexander Peltzer
ISMB Bioinfo Core Workshop NF-Core
Lightning talk introduction (5-8min) at ISMB 2018 Bioinfo Core Workshop, July 7th, 2:06PM at the Hyatt Regency Conference Hotel.
- 3,170



