X-Ray image segmentation
STFC/Durham University CDT in Data Intensive Science.
Carolina Cuesta-Lázaro
Arnau Quera-Bofarull
(Joseph Bullock)
Placement at IBEX Innovations Ltd.
Who are we?



2 months PhD placement at IBEX innovations, as part of the CDT program
Carolina / Arnau
Cosmology
Joe
Particle Physics
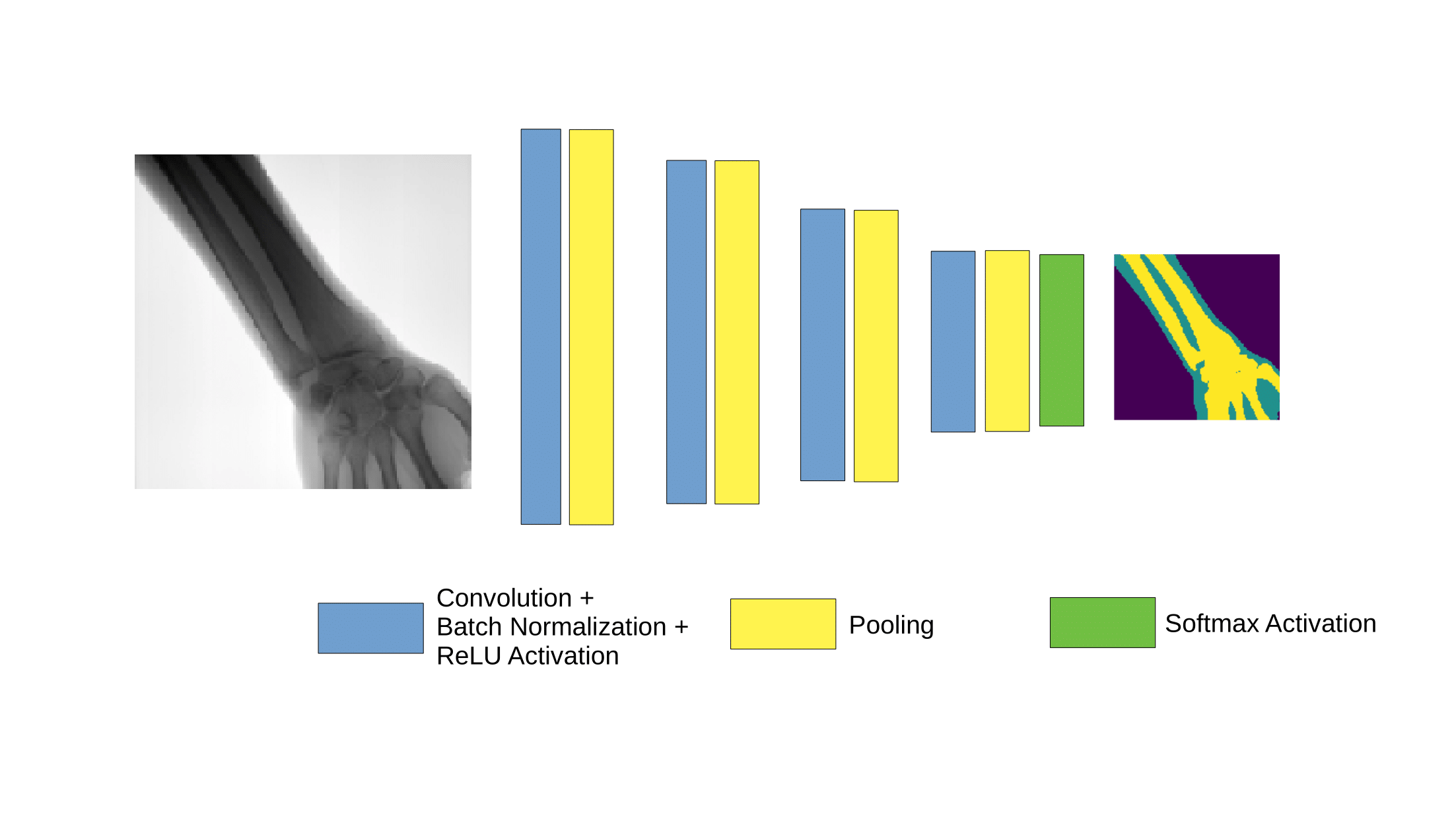
Detect bone and soft-tissue on X-Ray images
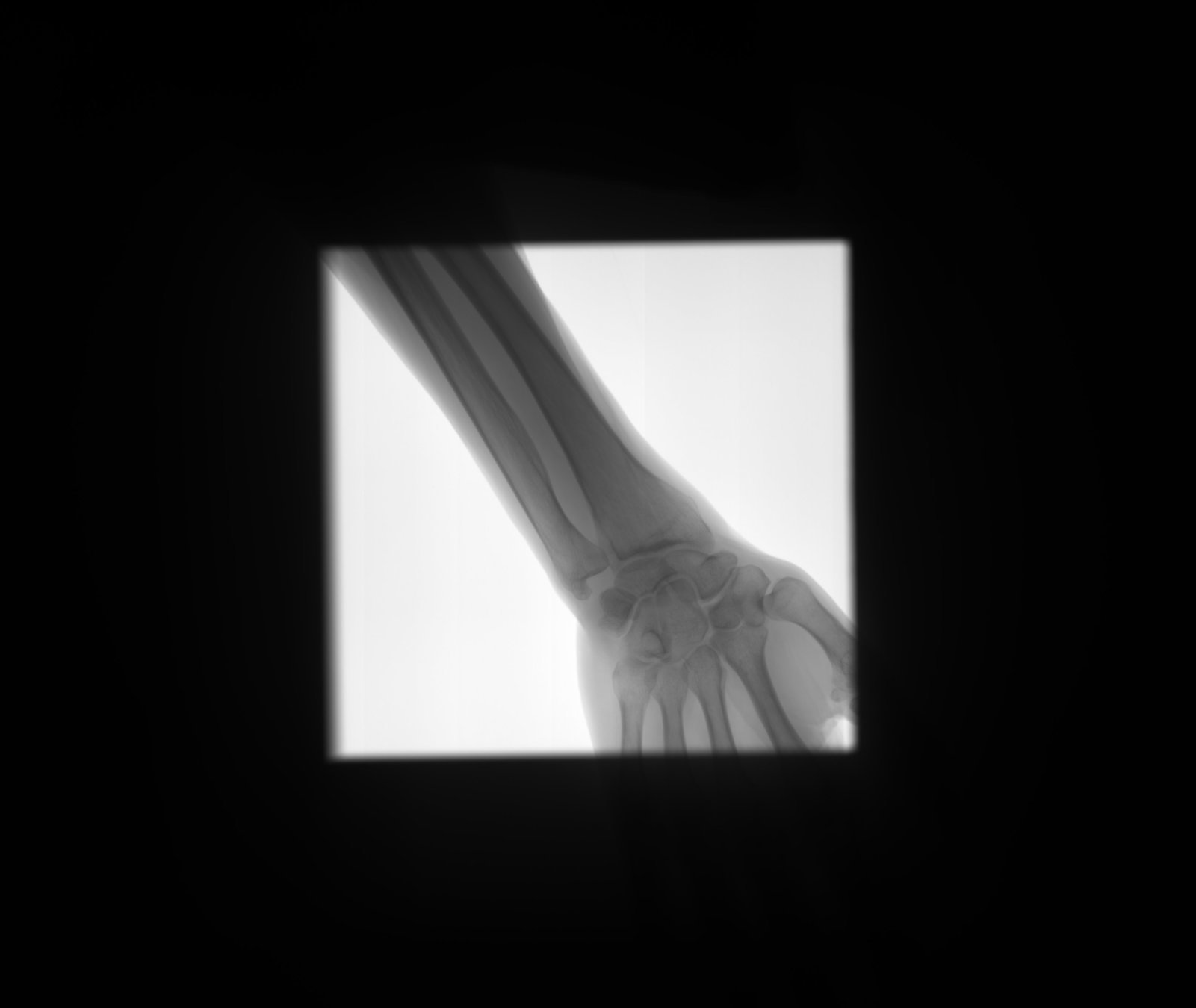
Detect
collimator

Segment

Open beam

Bone

Soft-tissue
Previous approaches
- Challenging problem due to varying brightness throughout the image.
- Usually done by detecting edges and shapes.
- High accuracy requires tuning of hyper parameters per image and body-part (not automated).
- Not well defined boundaries.
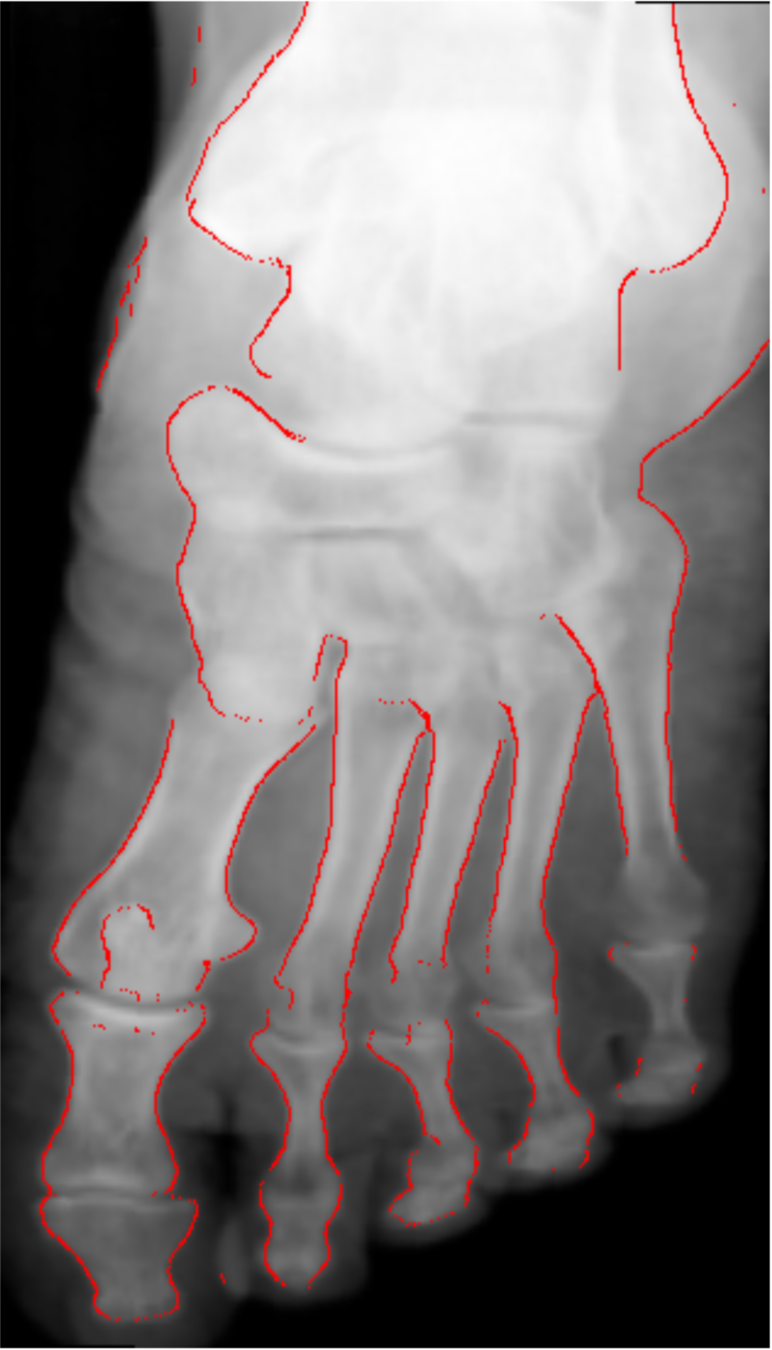
Kazeminia, S., Karimi, et al (2015)
Are there better features hidden in the data?
Extracts features from a high dimensional feature space, once trained on a particular dataset.
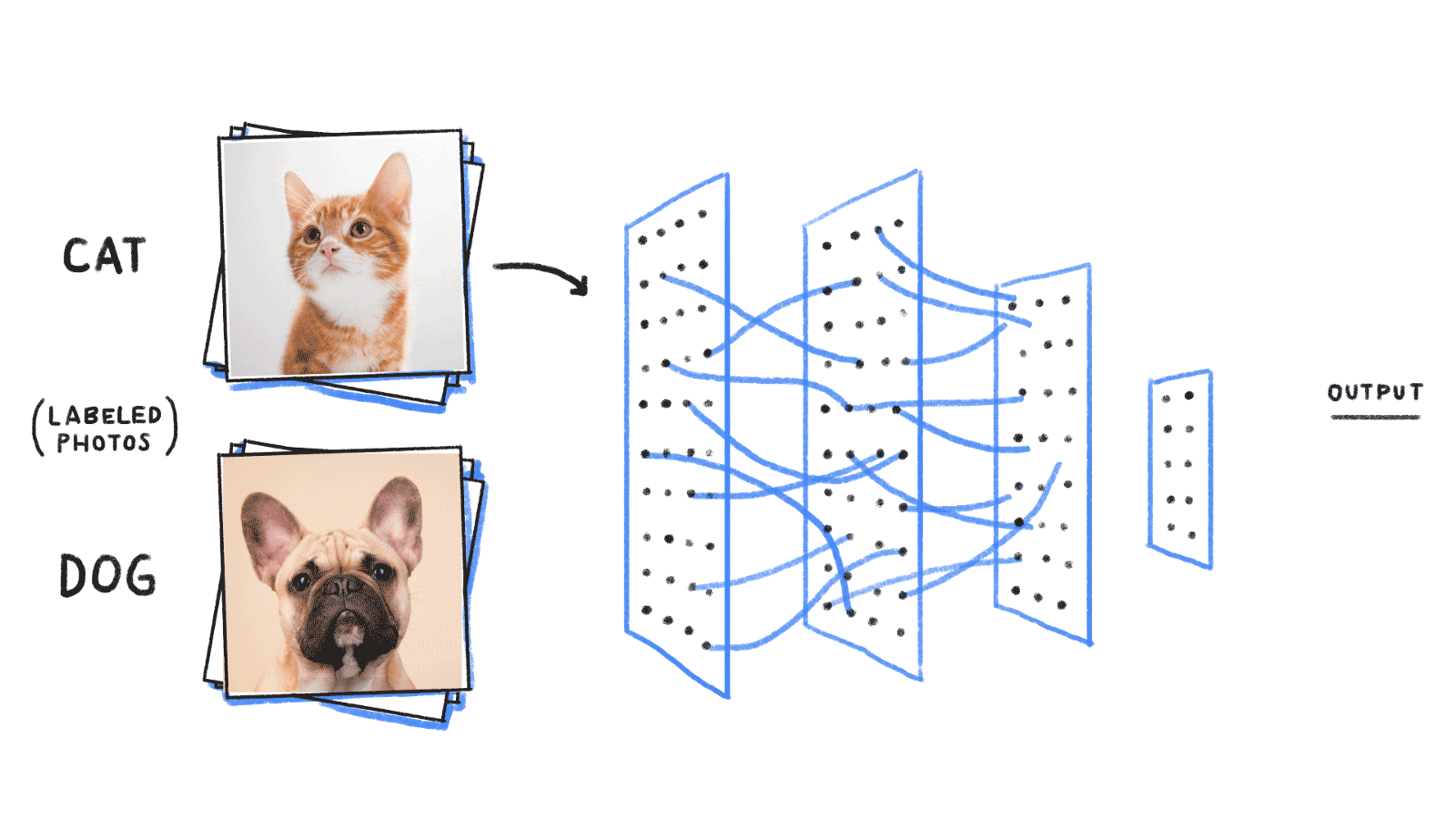
DEEP LEARNING

Deep Learning and image segmentation

http://cs229.stanford.edu/proj2017/final-reports/5241462.pdf
Švihlík et al (2014)
http://cs.swansea.ac.uk/~csadeline/projects_astro.html
Badrinarayanan et al (2015)
Challenges for segmentation with deep learning
Semantics (what)
MaxPooling

Translation Invariance
Location (where)
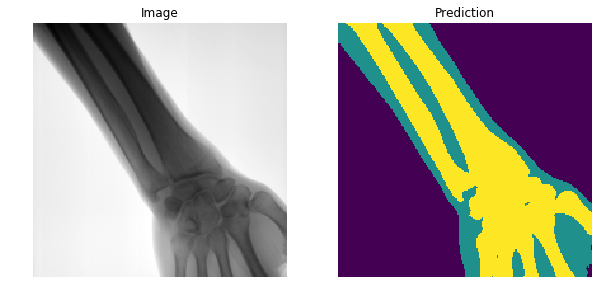
First attempt to segment
Receptive field grows very slowly !
Not tractable to learn long range correlations

Need pooling, but how to unpool?
Simple upsampling
| 1 | 0 |
|---|---|
| 2 | 3 |
| 1 | 1 | 0 | 0 |
|---|---|---|---|
| 1 | 1 | 0 | 0 |
| 2 | 2 | 3 | 3 |
| 2 | 2 | 3 | 3 |
Upsample
| 1 | 0 |
|---|---|
| 2 | 3 |
| 1 | 0 | 0 | 0 |
|---|---|---|---|
| 0.6 | 0.5 | 0 | 0 |
| 0.3 | 1 | 2 | 3 |
| 1.5 | 2 | 1.4 | 0.3 |
Use same indices that were pooled
Other layers
...
| 2 | 1 |
|---|---|
| 0.5 | 1.4 |
| 2 | 0 | 0 | 1 |
|---|---|---|---|
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 1.4 |
| 0 | 0.5 | 0 | 0 |
Transpose convolution
| 1 | 2 |
|---|---|
| 1 | 0 |
| 1 | 0 | 2 |
|---|---|---|
| 2 | 1 | 1 |
| 1 | 0 | 0 |
Learned filter
| 1 | 1 | 0 | 0 |
|---|---|---|---|
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
Stride 2 : Filter moves 2 pixels in the output for every pixel in the input
Feature map
| 1 | 1+4 | 2 | 2 |
|---|---|---|---|
| 0 | 0+2 | 0 | 0 |
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
Sum where overlap!
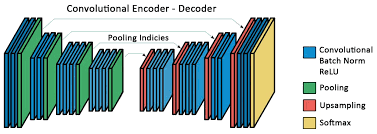
SegNet
- First Encoder - Decoder architecture for segmentation.
- Undo MaxPooling.
- The network has more than 15 Million free parameters.

Badrinarayanan et al (2015)
U-Net
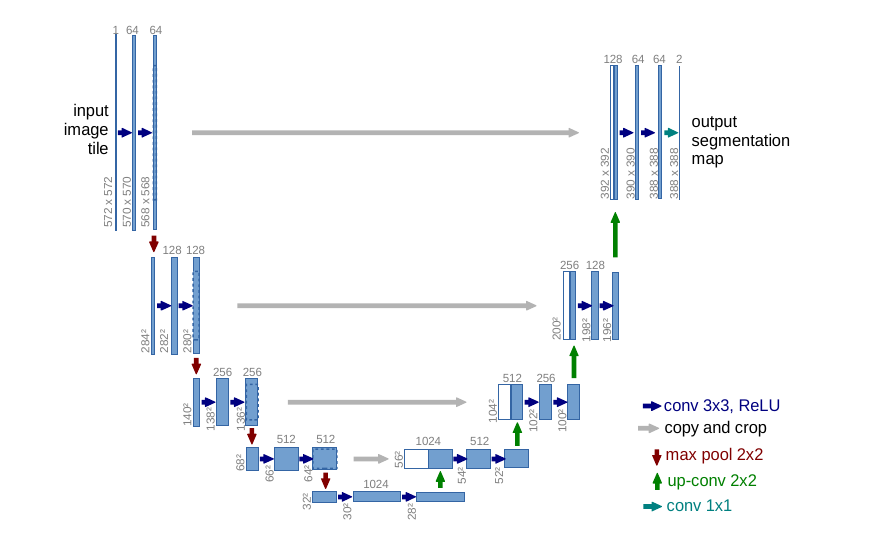
Skip connections to combine
information at different scales
Deconvolutions to
upsample
Up-weight the loss function
at the boundaries
Trained on 30 images for cell segmentation !
Ronneberger et al (2015)
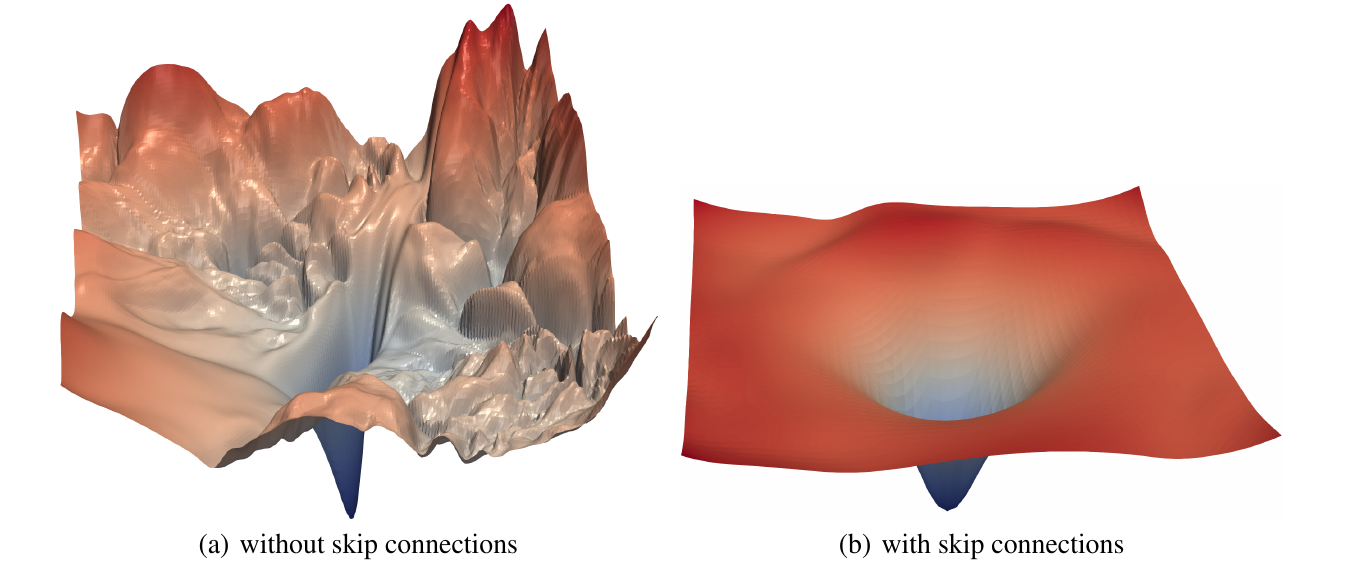
Skipping connections
Increases convexity and smoothness of loss function
DeepLab
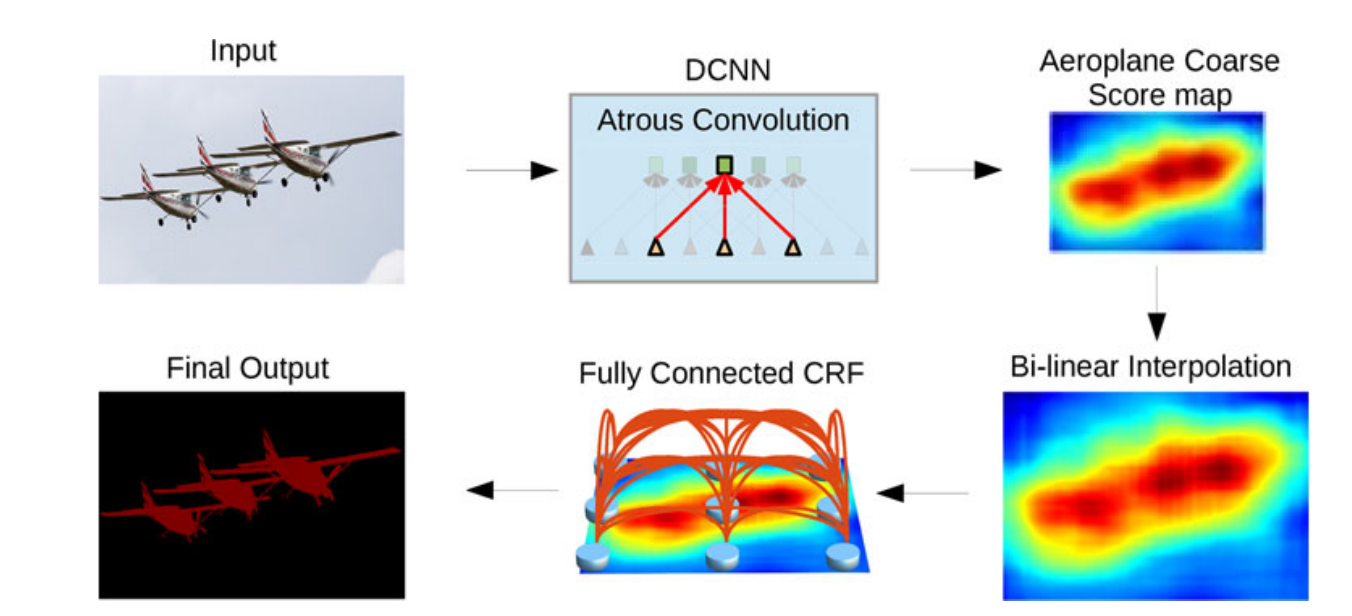
Pyramid of
Dilated Convolutions
Fast
Chen et al (2018)
Dilated Convolutions
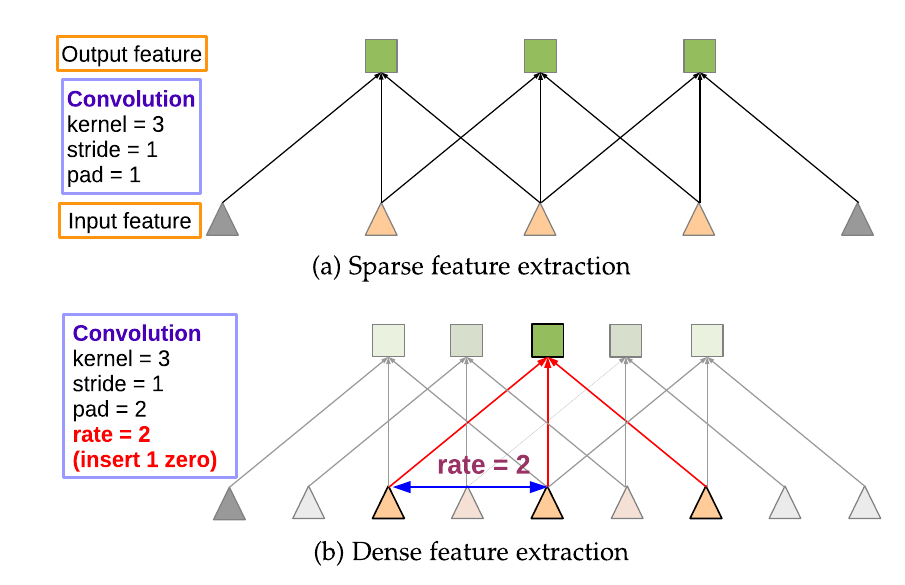

Chen et al (2018)
The Deep Learning Lego


Upsampling
Duplicate
Indices unpooling
Deconvolution
Interpolated
Preserve detail
Encoder - Decoder
Dilated Convolutions
Pyramid of convolutions
Architecture
Skip Connections
Asymmetric
Image credit : www.grabcad.com
The Road to XNet
Coursera
Dataset
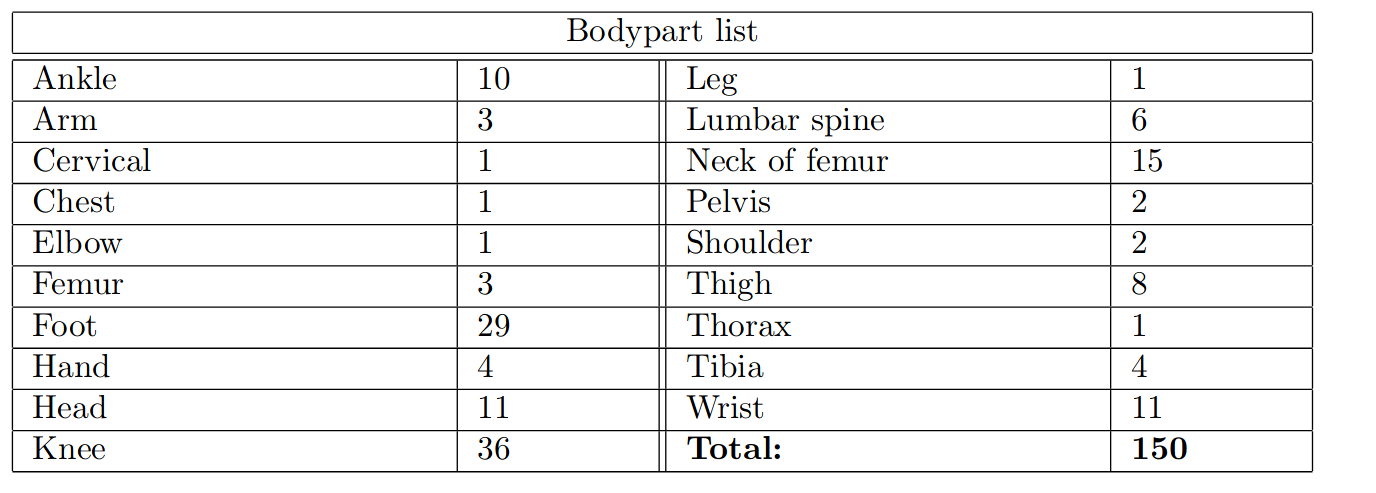
Small (compared to typical DL datasets) and unbalanced.
Dataset
How can we do Deep Learning with ~150 images?
-
Transfer Learning: Pre-trained network on different dataset.
Problems:- Not trivial to choose which layers to fine tune.
- Trained networks deal with very different data.
-
Data Augmentation: Add artificial data generated through transformations of the original dataset.
Problems:- Can add unwanted artifacts.
Data Augmentation
We generate new images by applying combinations of:
- Translations
- Rotations
- Shear transformations
- Cropping
- Elastic transformations
Splitting the dataset
We typically divide our dataset into three subsets:
- Training: 70% from categories with more than one sample.
Data augmentation -> Equal sample size for all categories.
Final size ~ 7000 images. - Validation: 15% from categories with more than one sample. Used to stop training and hyperparameter tuning.
- Test: 15% including categories with only one sample. Final network performance is evaluated in this set.
Network Architecture
First attempts focused on a very simplified SegNet model.
Underfitting
Network Architecture
Going deeper has limits ( limited image size, GPU memory bottleneck, overfitting).

Overfitting
XNet
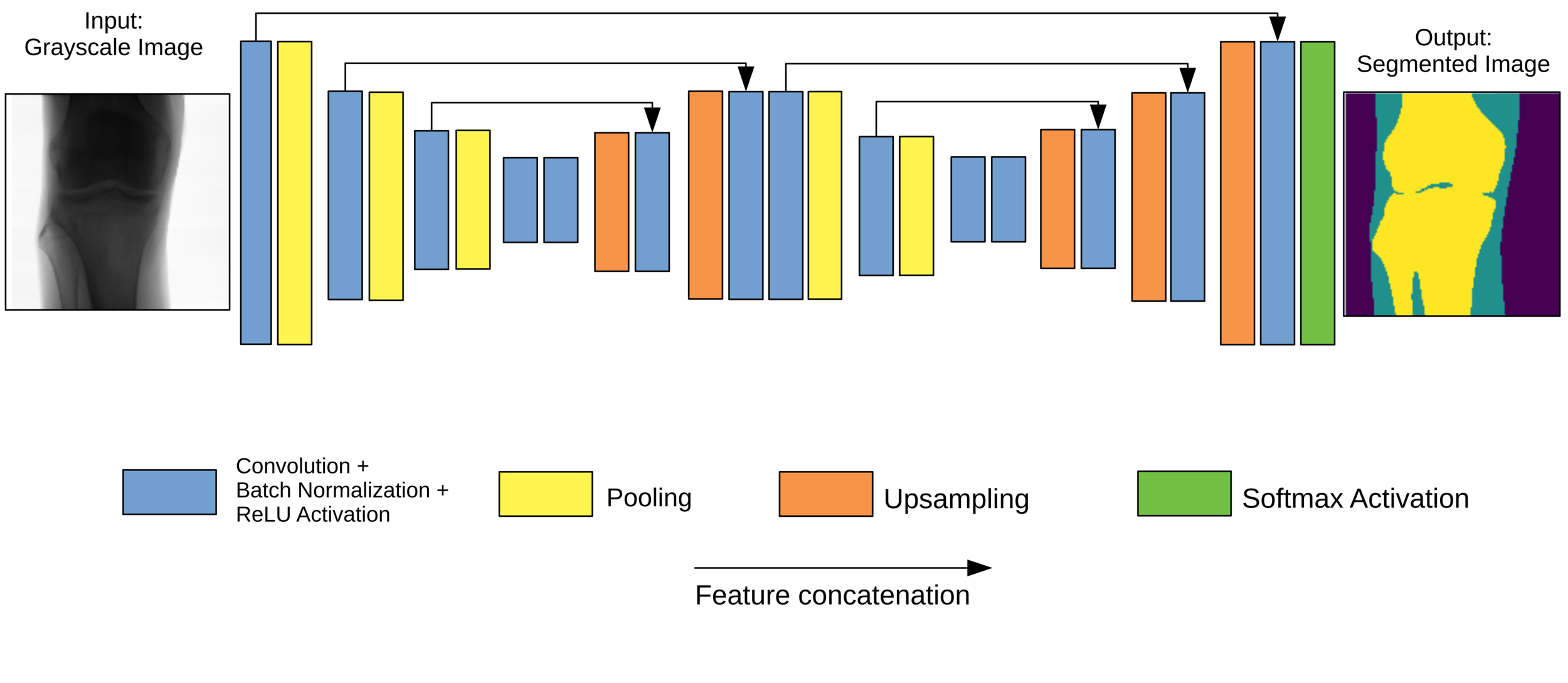
- Typical encoder-decoder architecture.
- W-shape for two feature extraction stages. Avoids resolution problems.
- Skip connections across levels.
- L2 regularisation at each convolutional layer.
Preventing overfitting
L2 regularisation
$$ L := - \sum_{i=1}^3 \color{royalblue}y_i \color{cadetblue}\log (f(x)_i) \color{black}+ \color{orange} \lambda \color{black}\sum_j \color{Maroon}\theta_j^2$$
Loss function = Cross-entropy + Regularisation
Regularisation hyperparameter $$\lambda = 5 \cdot 10^{-4}$$
Network parameters
Final model: XNet
Testing XNet
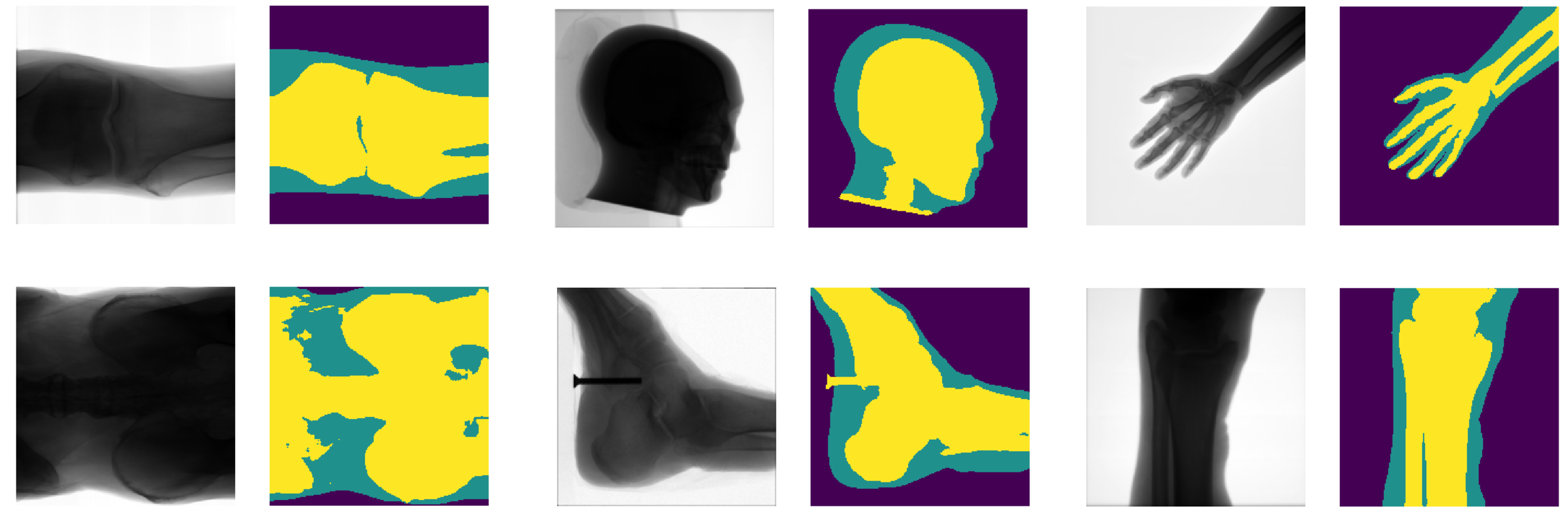
Danger! K-fold cross-validation needed.
Testing XNet
Metrics
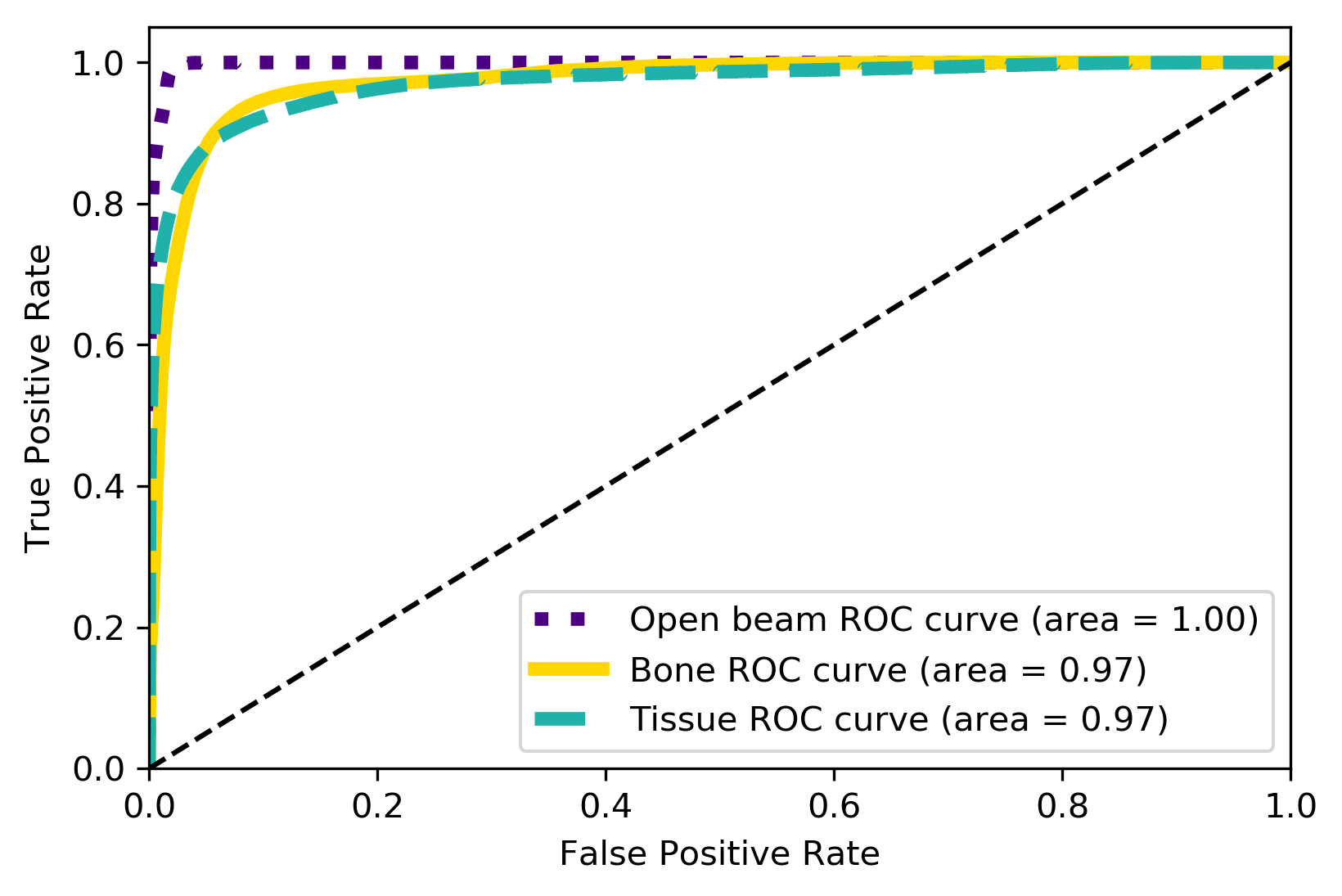
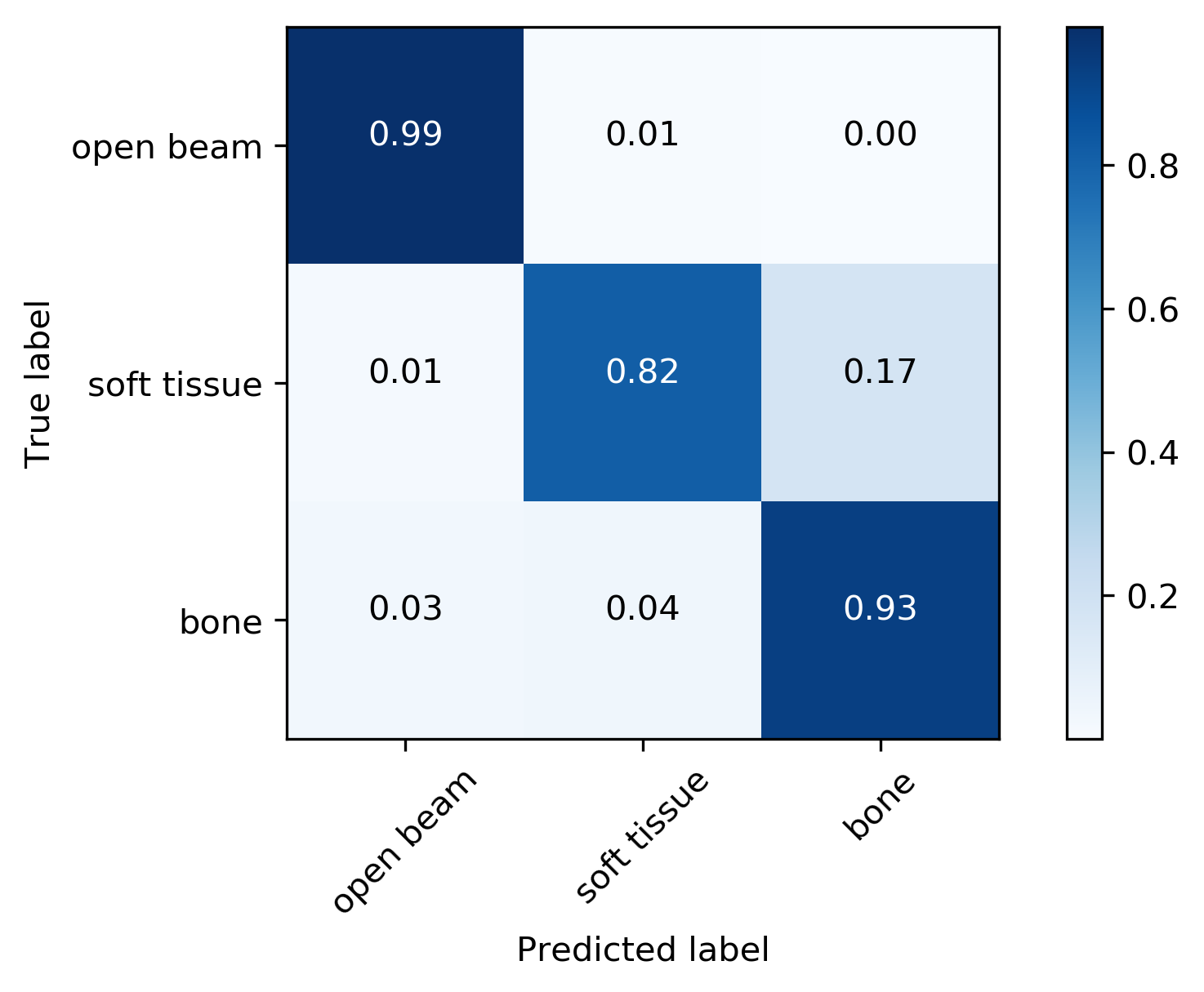
| Metrics | F1-Score | AUC | Accuracy | Confidence |
|---|---|---|---|---|
| Weighted Average | 0.92 | 0.98 | 92% | 97% |
Calibration?
TP Rate
FP Rate
True Label
Predicted Label
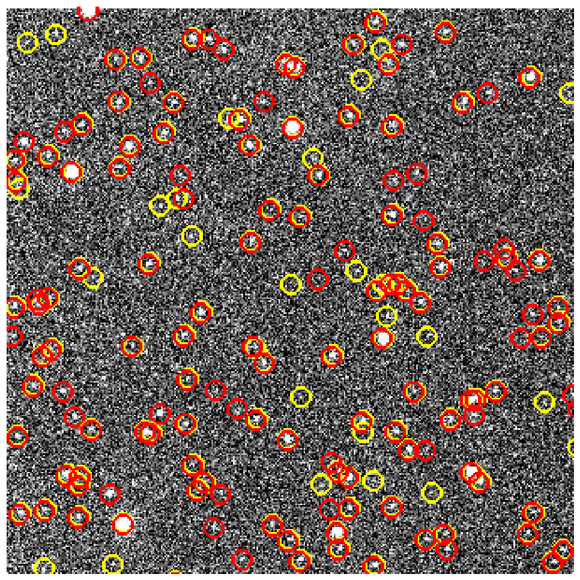
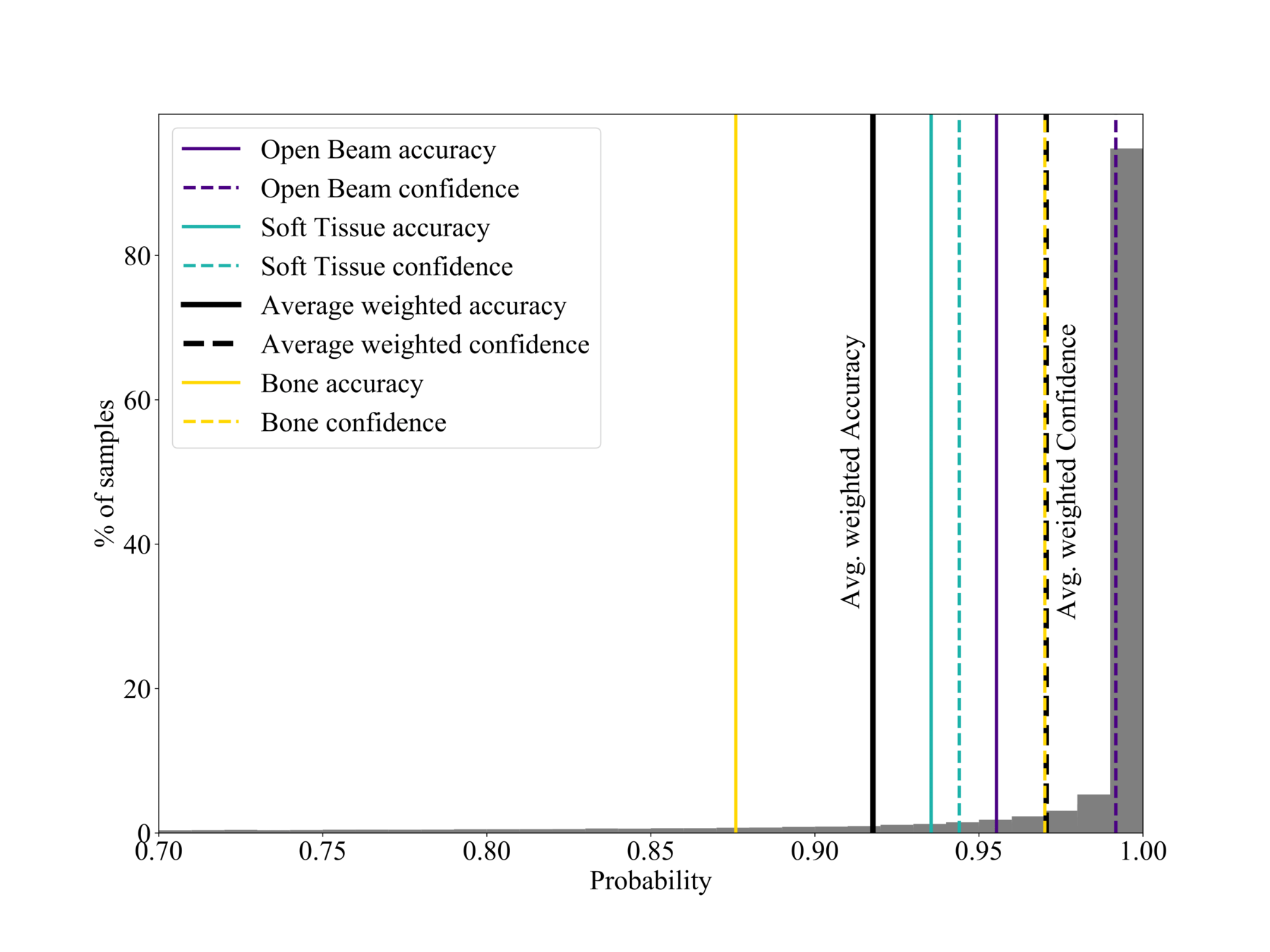
Calibration
Confidence vs Accuracy
- Moden CNNs often ill-calibrated (20% gap) (see Guo et al. (2017)).
- Reliable probability estimates important for post-processing.
- Network is a bit overconfident in the bone region, but overall acceptable (5% gap).
$$\text{Confidence}(X) = \frac{1}{| X|} \sum_{i \in X}p_i$$
Good calibration: Confidence ~ Accuracy
% of samples
Probability
Post-processing
Reducing soft tissue false positives.
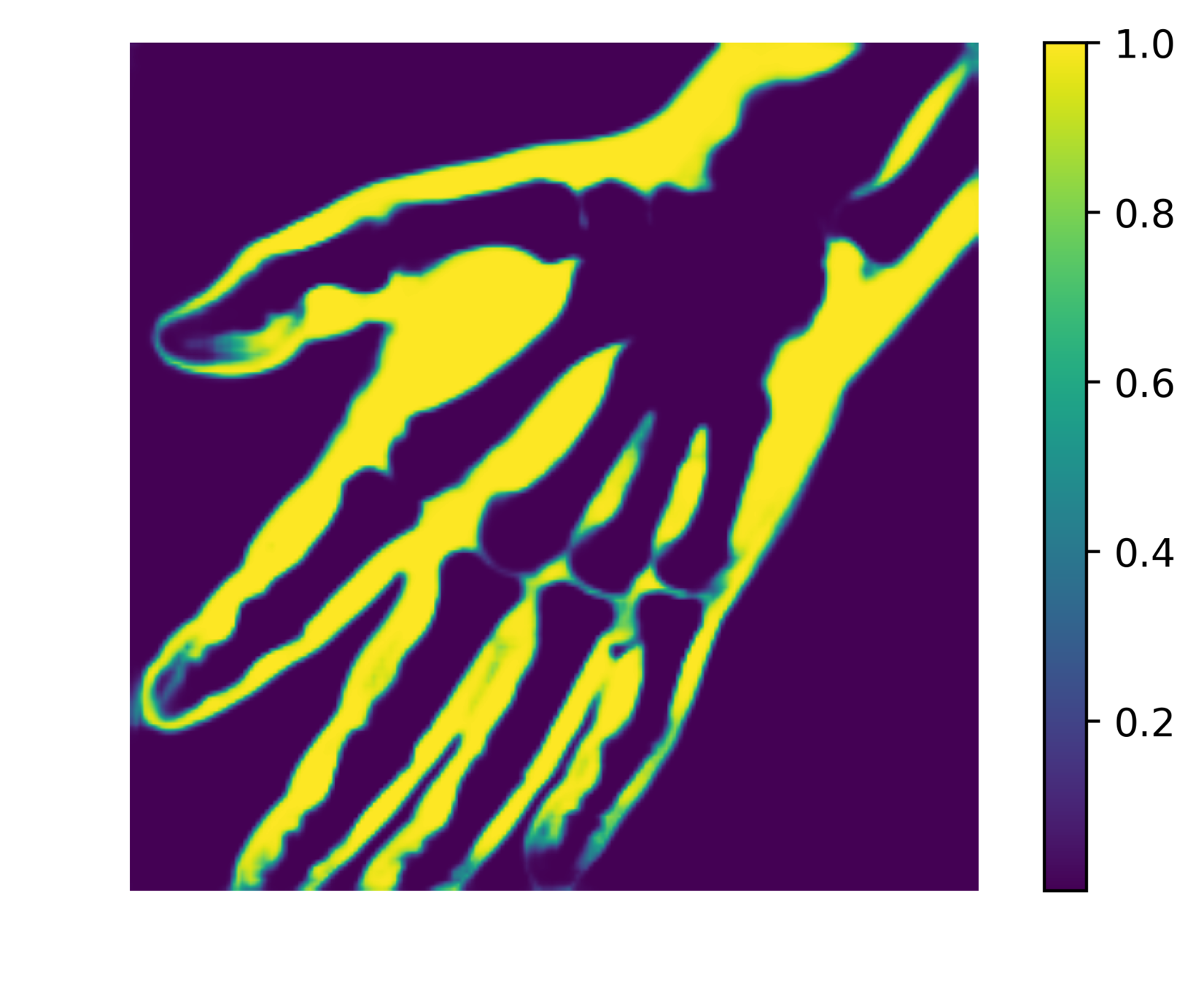
The network outputs 3 probability maps.
Soft tissue probability map
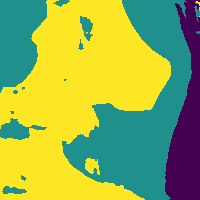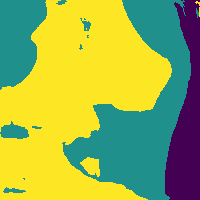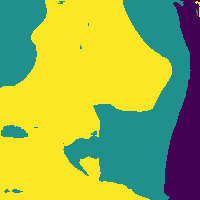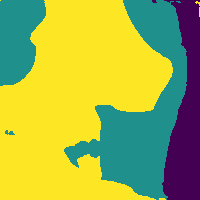
Apply threshold to probability map -> impose confidence level
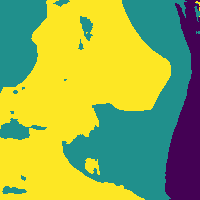
Probability
Comparison with other methods
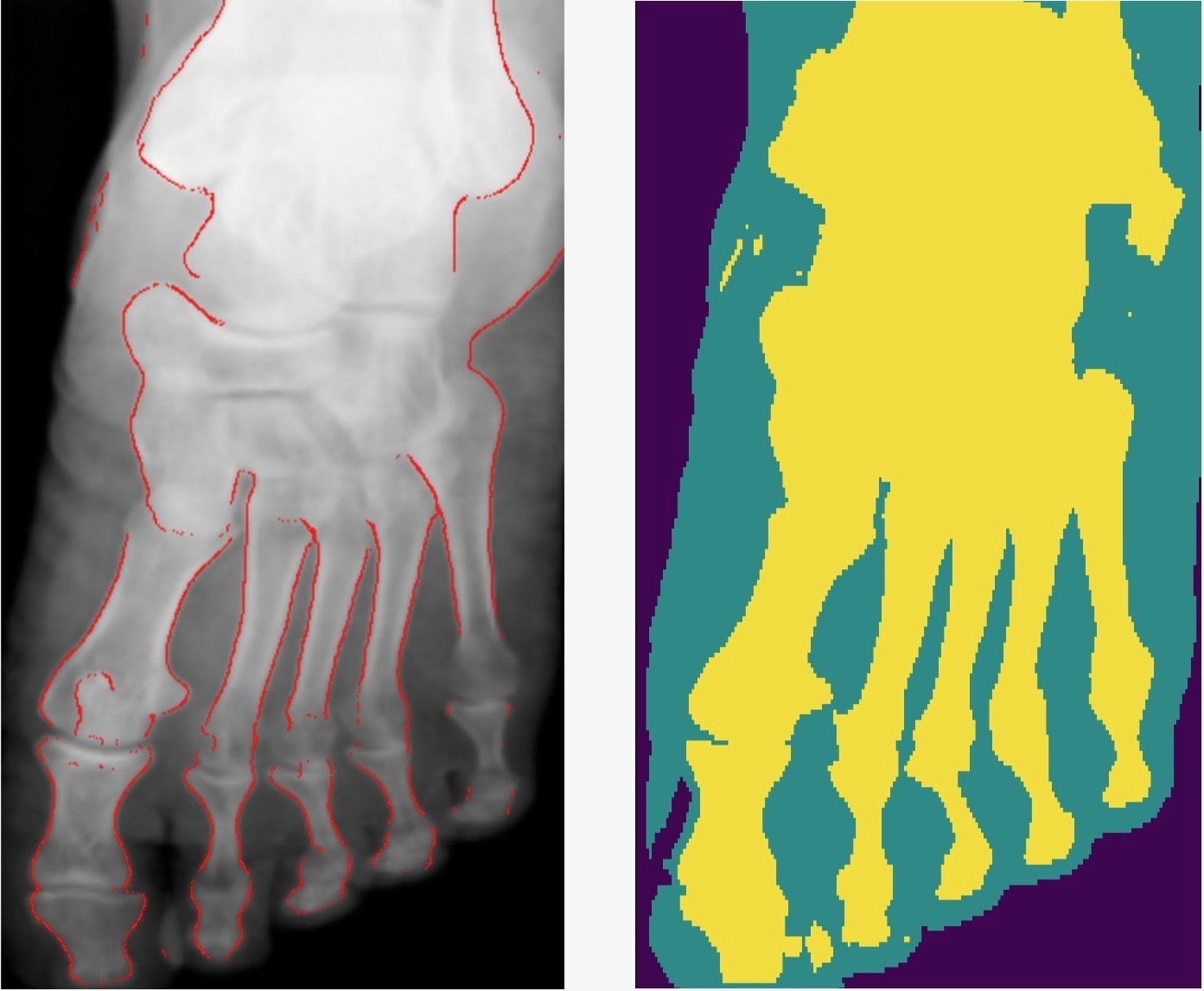
- Smooth connected boundaries.
- Better generalisation to different body parts (we do not have any frontal view of a foot in our dataset).
- More robust to noise.
- Well defined metrics to benchmark against.
Kazeminia, S., Karimi, et al (2015)
- The development process takes a long time due to hyperparameter tunning (50% of our internship time).
-
We used ~1000 GPU hours.
- 3x 4GB GPUS
- 1x 8GB GPU
- AWS 12GB GPU
Development process
Cryptocurrency Times
- Promising ML applications to medical imaging.
- Possible to train DL models with limited hardware and resources.
- Paper out arXiv:1812.00548v1, and presented at the SPIE Medical Imaging conference in San Diego. Best student paper awarded.
- Academia-industry collaboration exciting and rewarding for students.
Conclusions

X-Ray image segmentation
By arnauqb
X-Ray image segmentation
- 1,087



