Moving Code to Data
with workflow languages and containers
Context
Goals
My thesis project set out to test some things:
- Feasibility of OpenStack for research.
- Using common workflow language (CWL) on top of OpenStack.
- Using Docker containers to provide tools for easy researcher pipeline execution.
- Utilizing object storage on top of OpenStack to provide data for remote pipeline execution.
Data
-
Sharing of data is crucial in scientific community.
-
Increase of collaborative and reproducible work.
-
Data volumes are growing at substantial rates.
-
Storage devices are becoming considerably cheaper.
Cloud
Predominantly in the business field but reaching more and more into research.
More research oriented solutions popping up.
Still relatively expensive for high throughput.
More and more scientific data being held in cloud environments.
Justification
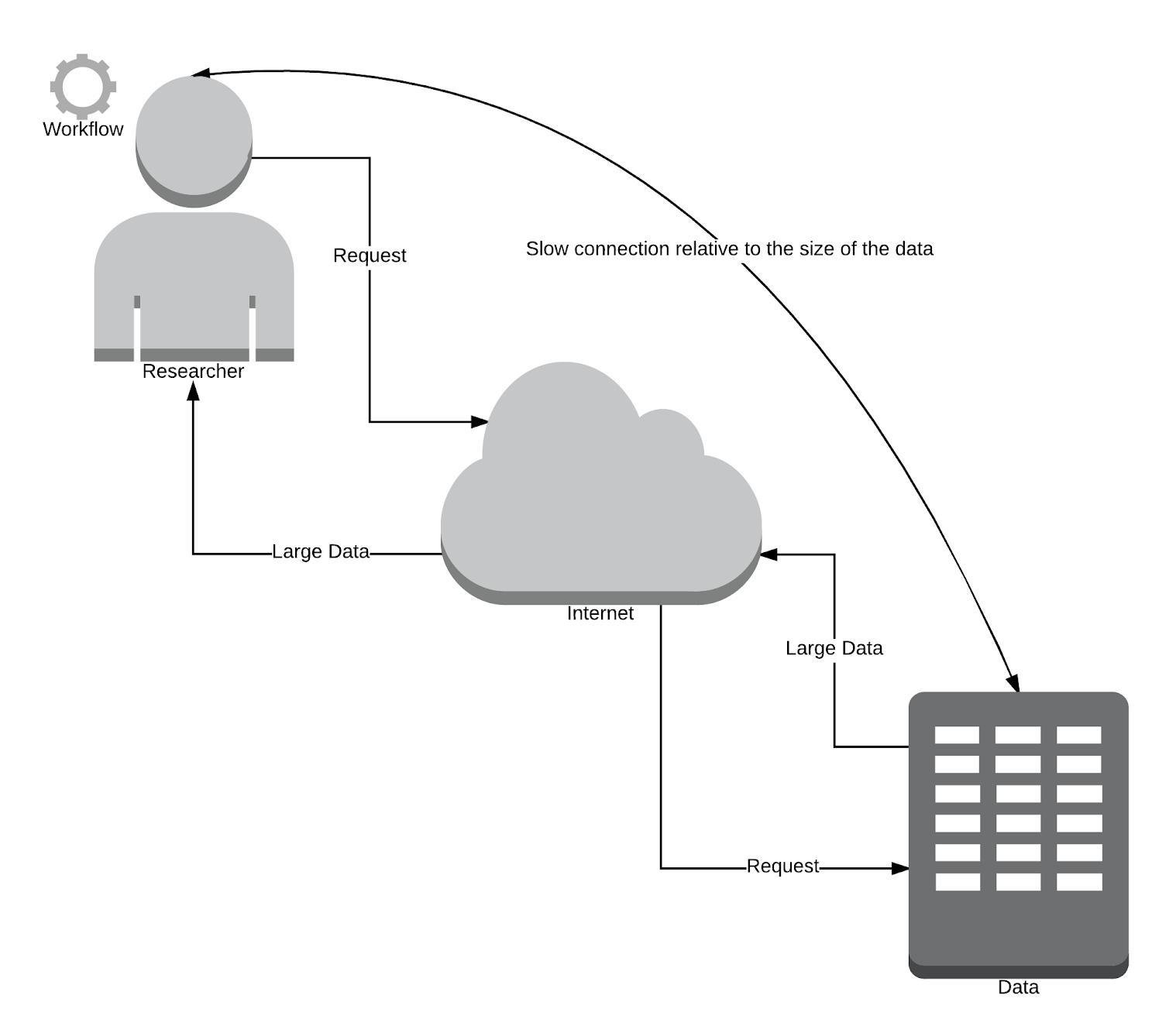
Traditional Way (vice versa)
Problems?
-
Data sizes grow faster than internet speeds (especially the case in developing countries).
-
Data governance issues with public clouds.
-
Researchers often need to know low level technical stuff, or work with someone who does.
Workflow Languages
Structured workflow definitions.
Reproducible code and workflows.
Easier sharing.
Many growing in support for containerization.
Easier to support from a systems perspective.



Masters Thesis
Assumptions
-
Data exists at the institute.
-
Institute provides a cloud environment.
OpenStack
-
Self-hosted cloud.
-
Open source and free (if not using commercial offering).
-
Gaining traction as an alternative to using commercial products.
-
South Africa has projects currently using or looking at utilizing OpenStack:
-
ARC - Now called SADIRC.
-
IDIA.
-

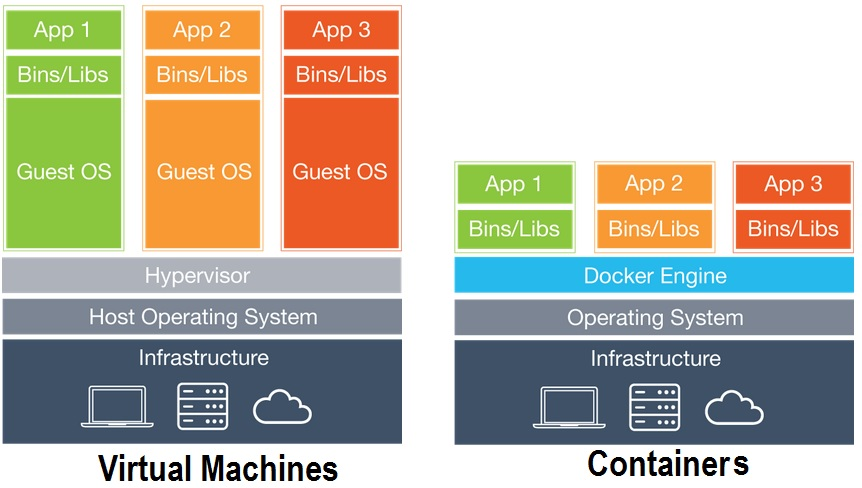
Software Containers
-
Light-weight alternative to virtualization.
-
Reproducible software environments.
-
Easily shareable code.
-
Write once, run anywhere.
-
Some container systems are build specifically for science.

Quick Overview of Vitual Machines vs. Containers
(docker.io)
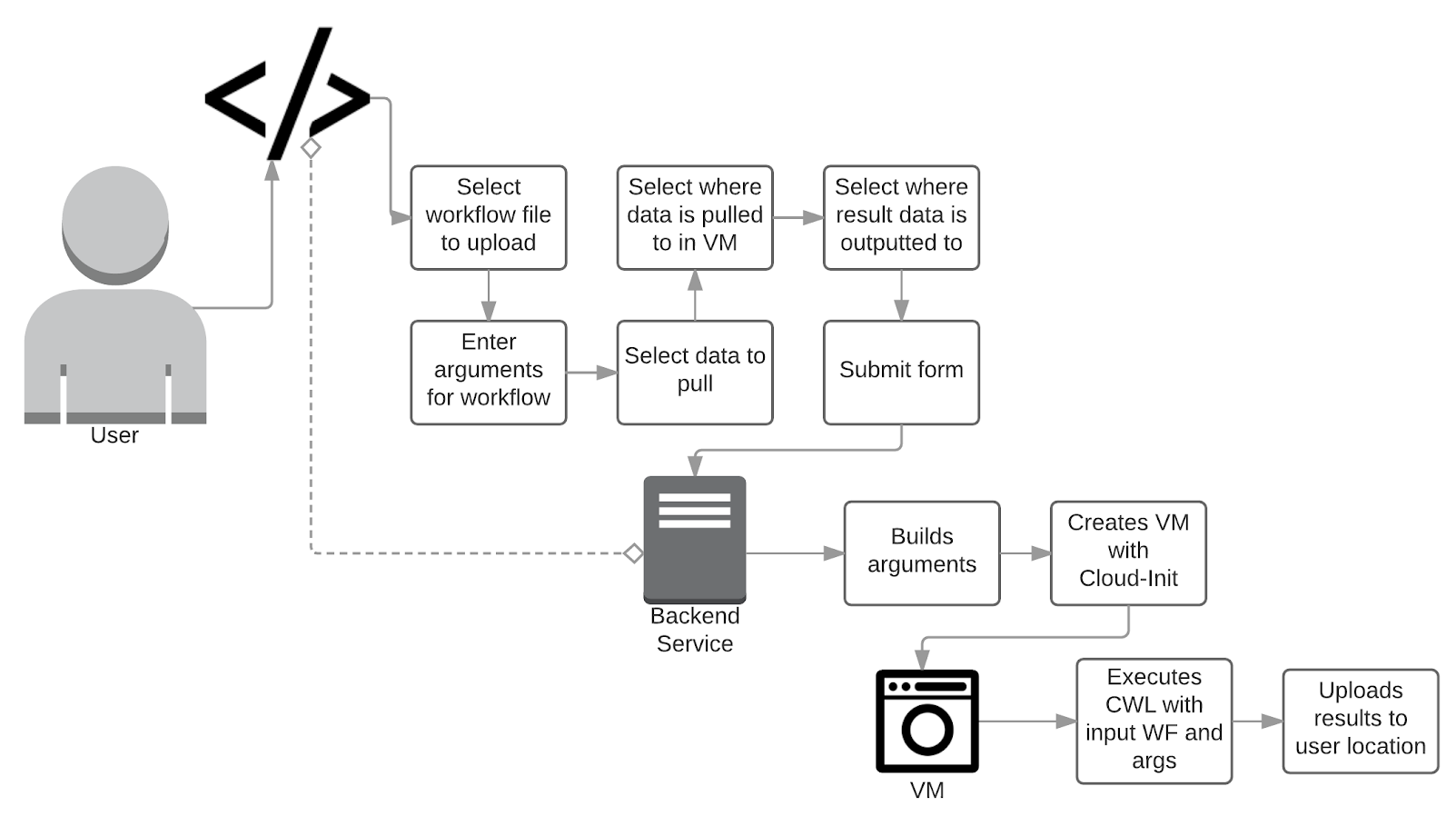
Proposed Solution
-
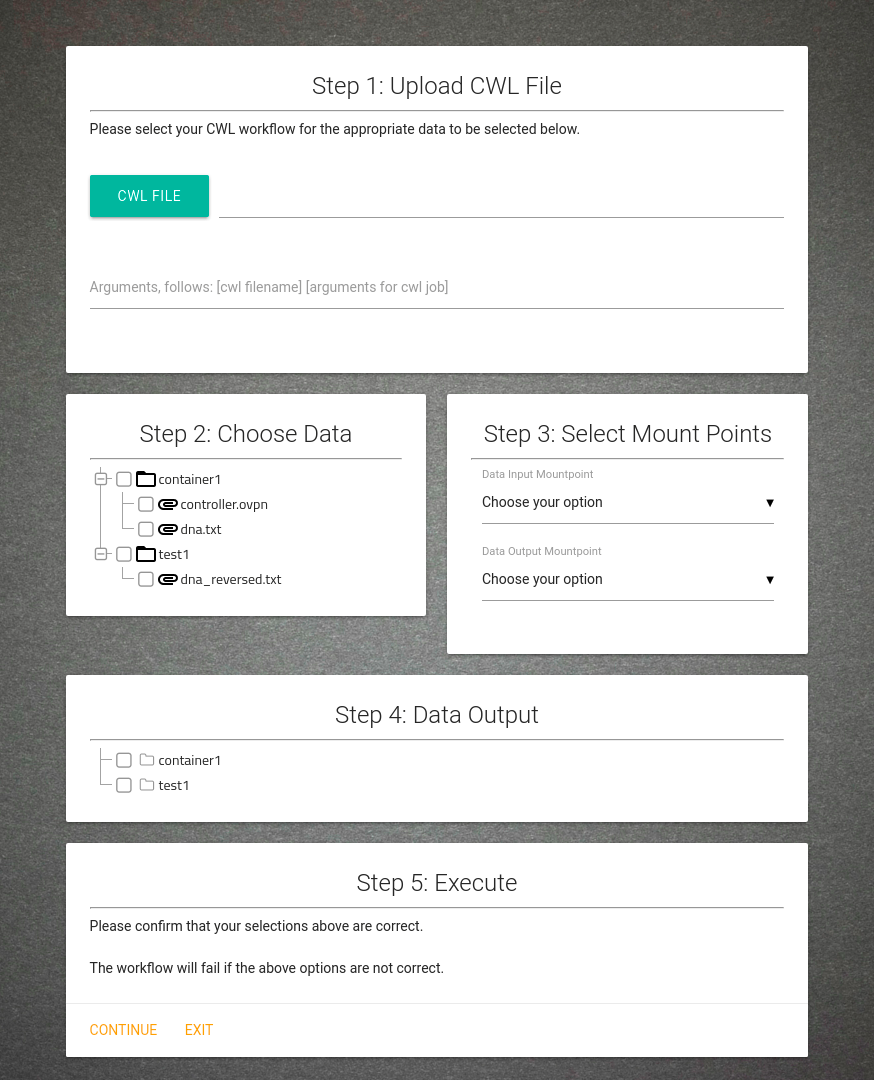
Submit workflow definitions.
-
Specify run-time arguments for workflow execution.
-
Select data from list of data authorized for their use.
-
Select where the resultant data is sent to.
-
Use the system without knowledge of back-end cloud infrastructure or use of said cloud environment.
-
Autonomously deploy containers from workflow definition and execute workflow.
Implementation
#!/usr/bin/env cwl-runner
cwlVersion: v1.0
requirements:
- class: DockerRequirement
dockerPull: quay.io/ncigdc/fastqc:1
- class: InlineJavascriptRequirement
class: CommandLineTool
inputs:
- id: adapters
type: ["null", File]
inputBinding:
prefix: --adapters
- id: casava
type: ["null", boolean]
default: false
inputBinding:
prefix: --casava
- id: contaminants
type: ["null", File]
inputBinding:
prefix: --contaminants
- id: dir
type: string
default: .
inputBinding:
prefix: --dir
- id: extract
type: boolean
default: false
inputBinding:
prefix: --extract
- id: format
type: string
default: fastq
inputBinding:
prefix: --format
- id: INPUT
type: File
format: "edam:format_2182"
inputBinding:
position: 99
- id: kmers
type: ["null", File]
inputBinding:
prefix: --kmers
- id: limits
type: ["null", File]
inputBinding:
prefix: --limits
- id: nano
type: boolean
default: false
inputBinding:
prefix: --nano
- id: noextract
type: boolean
default: true
inputBinding:
prefix: --noextract
- id: nofilter
type: boolean
default: false
inputBinding:
prefix: --nofilter
- id: nogroup
type: boolean
default: false
inputBinding:
prefix: --nogroup
- id: outdir
type: string
default: .
inputBinding:
prefix: --outdir
- id: quiet
type: boolean
default: false
inputBinding:
prefix: --quiet
- id: threads
type: int
default: 1
inputBinding:
prefix: --threads
outputs:
- id: OUTPUT
type: File
outputBinding:
glob: |
${
function endsWith(str, suffix) {
return str.indexOf(suffix, str.length - suffix.length) !== -1;
}
var filename = inputs.INPUT.nameroot;
if ( endsWith(filename, '.fq') ) {
var nameroot = filename.slice(0,-3);
}
else if ( endsWith(filename, '.fastq') ) {
var nameroot = filename.slice(0,-6);
}
else {
var nameroot = filename;
}
var output = nameroot +"_fastqc.zip";
return output
}
baseCommand: [/usr/local/FastQC/fastqc]
-
Ran a CWL file with a Dockerized Fastqc tool.
-
Short read data file from NCBI was used as example (NA12878).
-
Input data was 2.77 GB, preloaded on cloud environment.
-
Resultant data was 650 KB, generated on cloud environment.
-
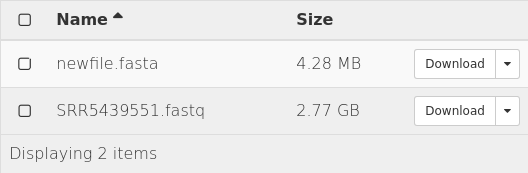
Result

test data
result data
Findings


This project was a simple proof of concept
This concept has a bright future
Thanks
Prof. A Christoffels, Peter van Heusden
UWC Astrophysics and ICS departments



Reach Me At:
eugene@sanbi.ac.za
https://themeanti.me
GA2018
By Eugene de Beste
GA2018
- 770


