Student Experience
STFC/Durham University CDT in Data Intensive Science.
Carolina Cuesta-Lázaro
Arnau Quera-Bofarull
(Joseph Bullock)
Placement at IBEX Innovations Ltd.
Who are we?



2 months team project at IBEX innovations
Carolina / Arnau
Cosmology
Joe
Particle Physics
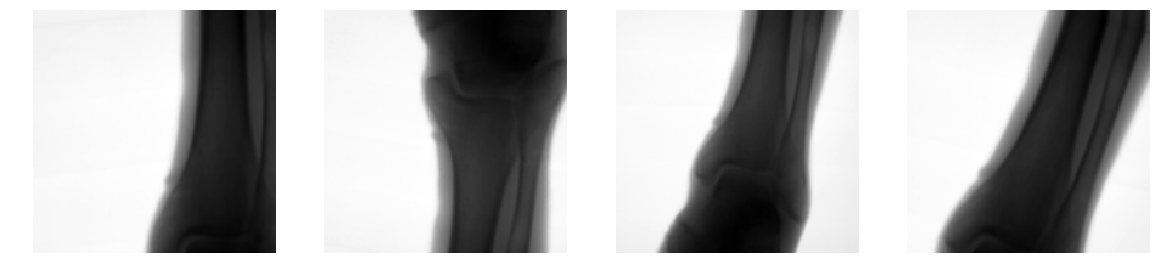
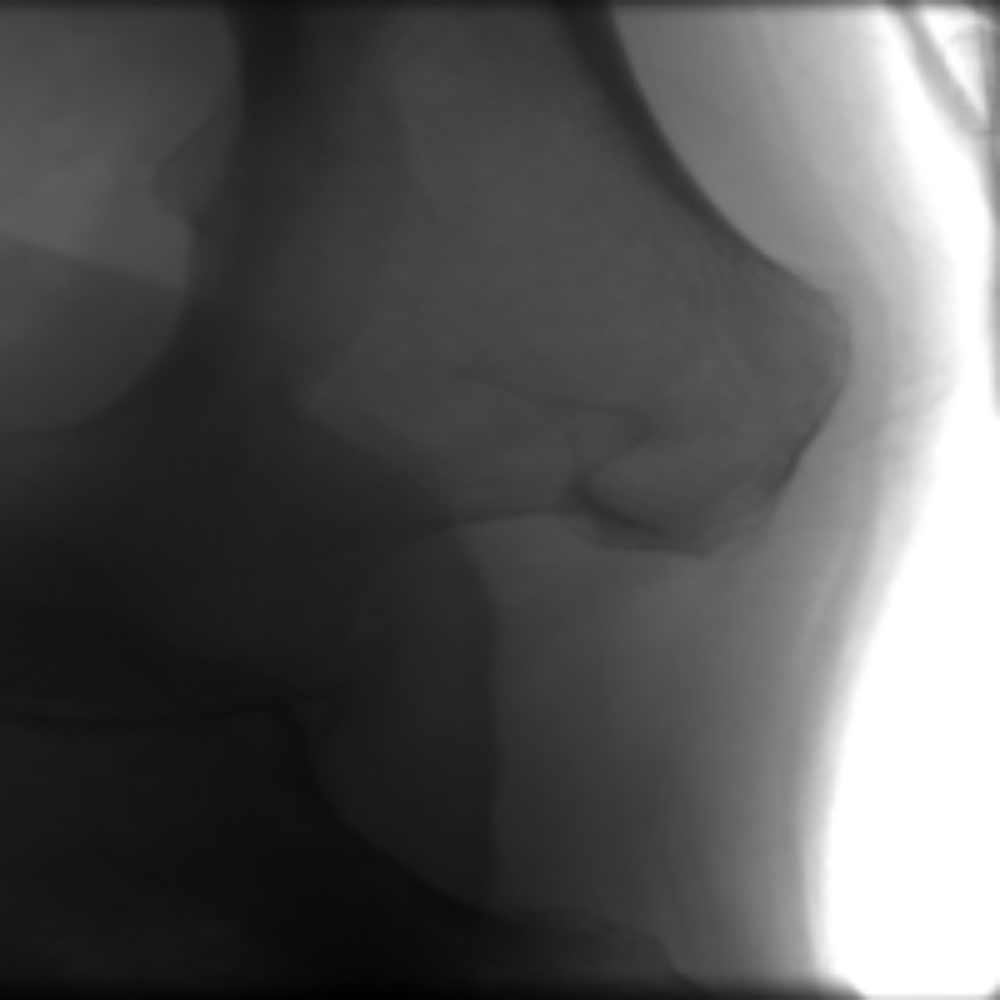
Detect bone and soft-tissue on X-Ray images
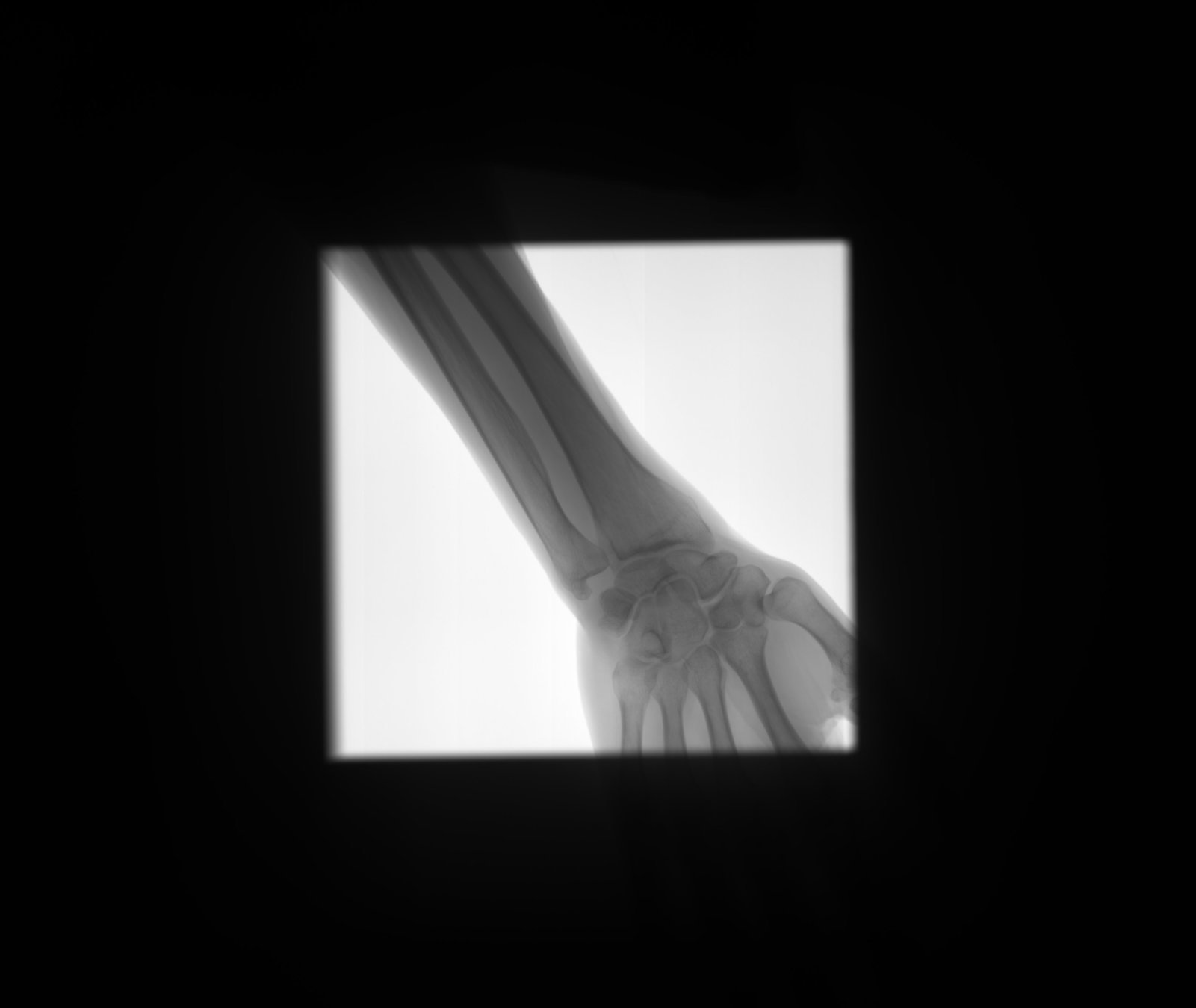
Detect
collimator

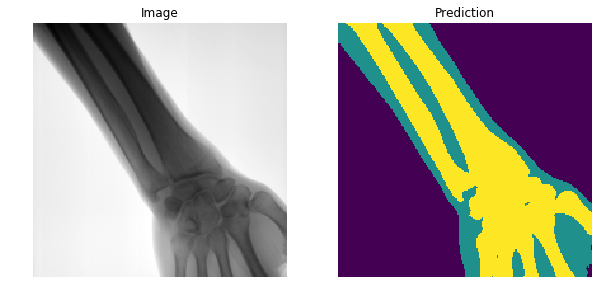
Segment

Open beam

Bone

Soft-tissue
Previous approaches
- Challenging problem due to varying brightness throughout the image.
- Usually done by detecting edges and shapes.
- High accuracy requires tuning of hyper parameters per image and body-part (not automated).
- Not well defined boundaries.
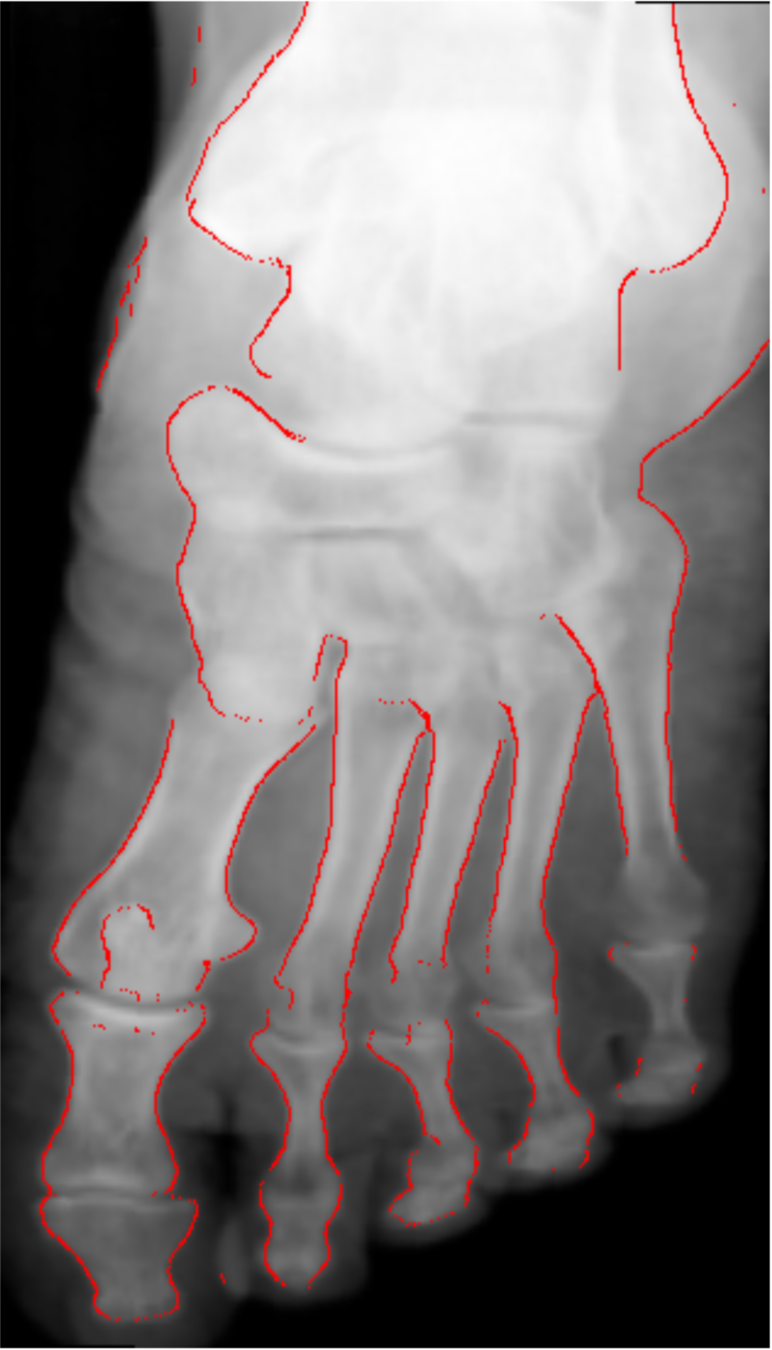
Kazeminia, S., Karimi, et al (2015)
Are there better features hidden in the data?
Extracts features from a high dimensional feature space, once trained on a particular dataset.
DEEP LEARNING

What is training?
Luminosity
Size
Colour
Galaxy
Star
x 2

LOSS = GENERATED OUTPUT - ACTUAL OUTPUT
Galaxy
Star
x 4
Deep learning on images
Credit : https://www.pnas.org/content/116/4/1074
With the right data, the network will find the right features

With wrong data ....
Does it really work?
SegNet
- The network has more than 15 Million free parameters.
- To find the values of the parameters that produce the correct segmentation, it has been trained on 1.3 Million images.

Could it solve our problem?
150 labeled images.
Hardware limitations (memory, training time...).
We need a fast network, easier to re-train as we get more images.
CONS
PROS
Could work for different detectors (different noise).
Generalize to different body-parts.
Well defined boundaries between regions.
Could be improved through more training.
The Dataset
- Small, ~150 images
- Unbalanced
Solution:
Artificially augment the dataset by transforming original images.
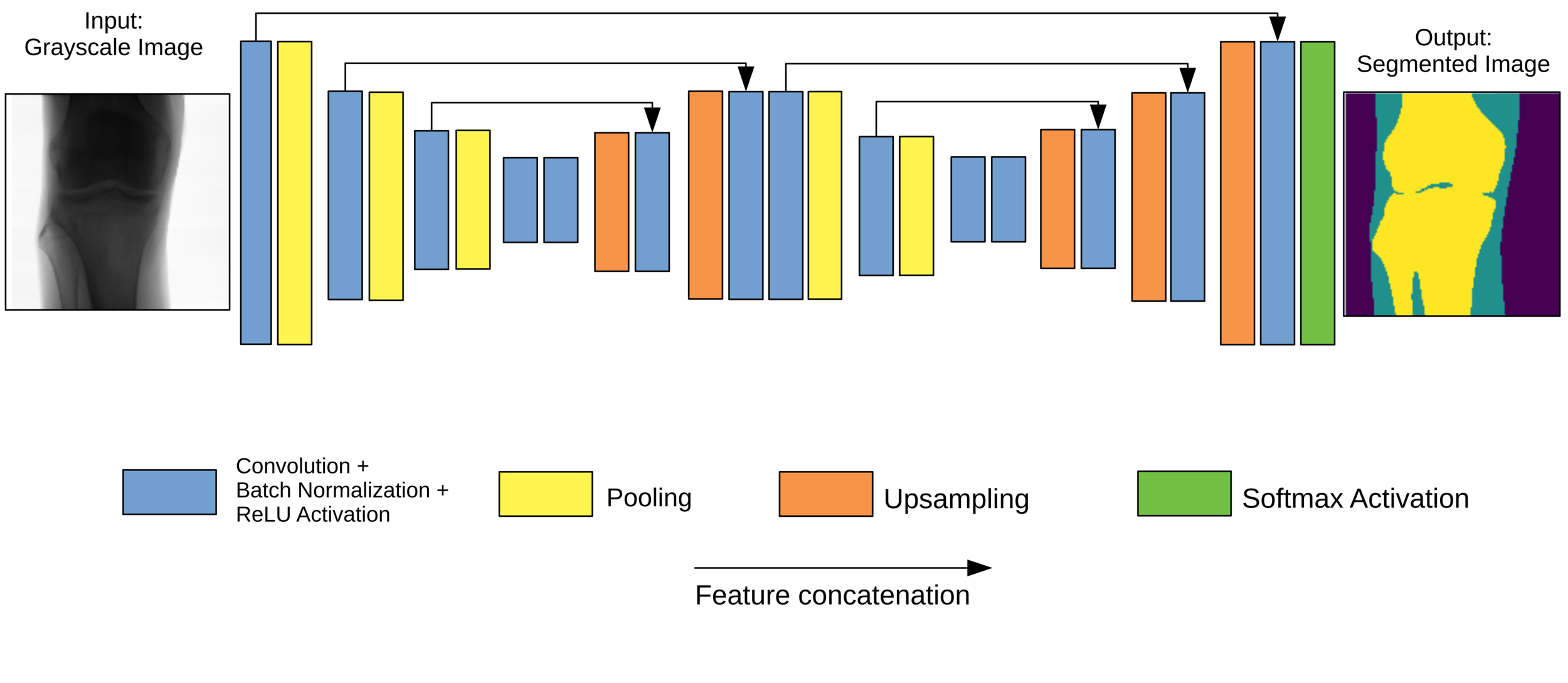
Network Architecture
Original Architecture based on SegNet but much fewer parameters.
How did we get here?
Training
Dataset
Test set (15%)
Cross-validation set (15%)
Training set (70%)
- The network minimises the loss function on the training set.
- We check how well the network is doing with the cross-validation set.
- The final network accuracy is the performance on the test set.
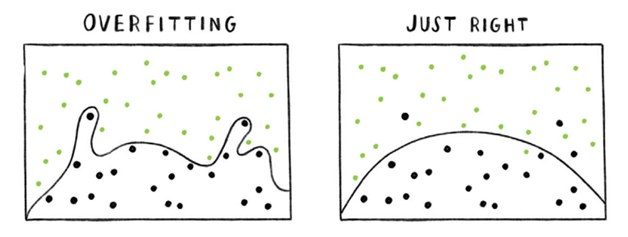
Debugging a network
Simple Network, low number of parameters.
Underfitting
Debugging a network
Deeper Network, high number of parameters.

Overfitting
Dealing with overfitting
Ways to reduce overfitting
- Increase the dataset.
- Reduce network complexity.
- Regularisation.
Idea:
Penalise the network if it uses too many parameters to fit the data.
Credit: www.kdnuggets.com
Final model: XNet
Post-processing
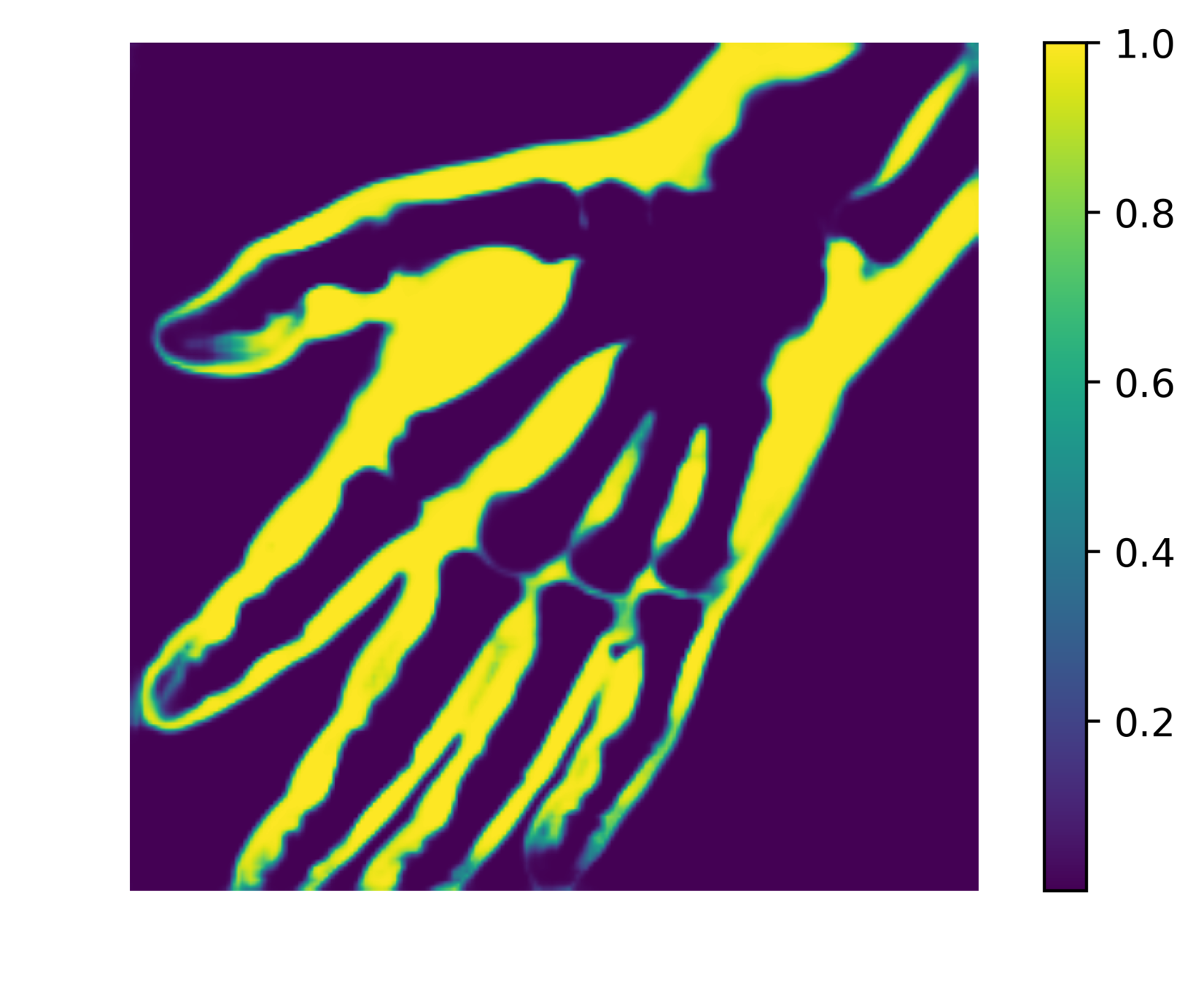
The network outputs 3 probability maps.
Soft tissue probability map
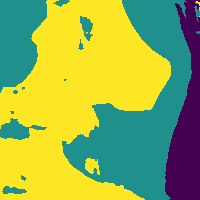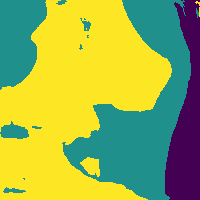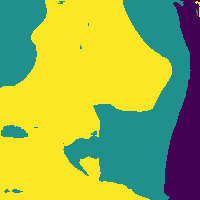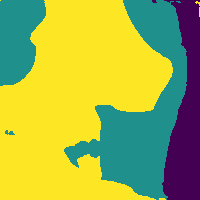
We can reduce the number of false positives by making a probability cut to the map.
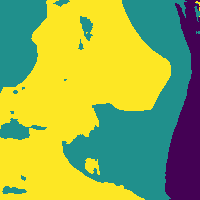
Probability
Results
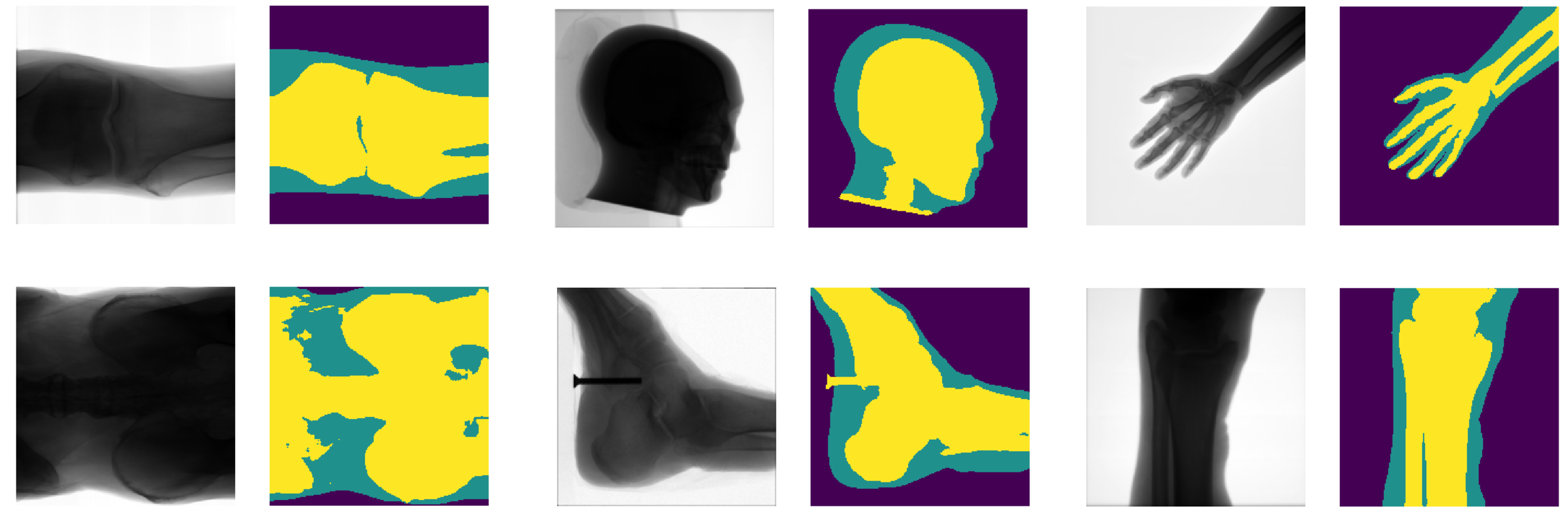
- Generalises well even for unseen categories!
- Overall accuracy on test set: 92%
- Soft tissue TP/FP rate: 82% / 4%
Comparison with other methods
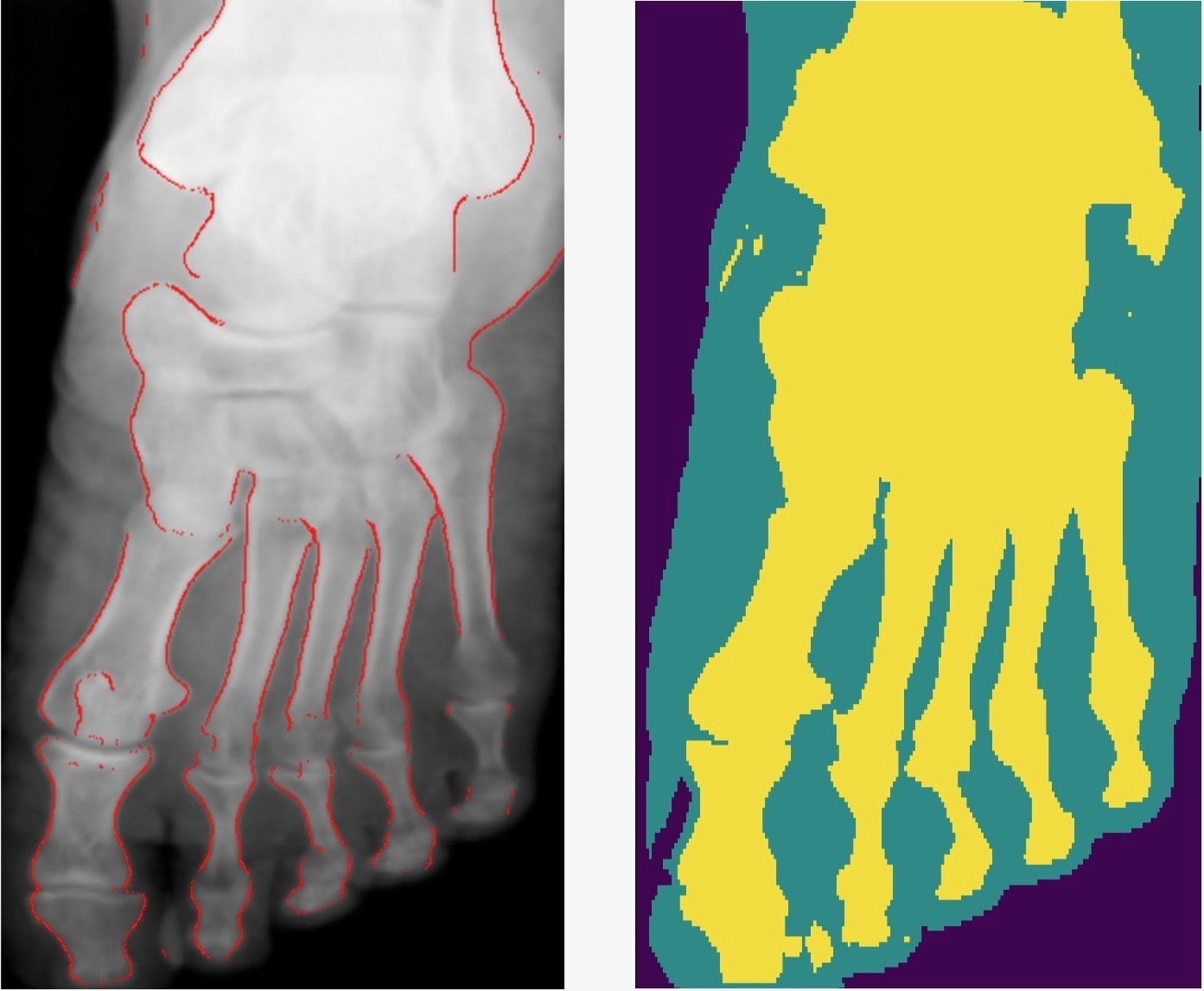
- Smooth connected boundaries.
- Better generalisation to different body parts (we do not have any frontal view of a foot in our dataset).
- More robust to noise.
- Well defined metrics to benchmark against.
- The development process takes a long time due to hyperparameter tunning (50% of our internship time).
- We used ~1000 GPU hours.
- 3x 4GB GPUS
- 1x 8GB GPU
- AWS 12GB GPU
Development process
Coursera
Cryptocurrency Times
- Promising ML applications to medical imaging.
- Possible to train ML models with limited hardware and resources.
- Knowledge of building and deploying a machine learning product in an industrial setting.
- XNet Paper is on the arXiv:1812.00548v1, and will be presented in the upcoming SPIE Medical Imaging conference in San Diego.
Conclusions
Learning outcomes
beyond-the-lab
By carol cuesta
beyond-the-lab
- 528



