Differential innervation of GPi → LHb and LHA → LHb projection neurons
Rania Tzortzi
Meletis Lab
Meletis lab meeting
4th July 2016
rania@ki.se


Differential innervation of GPi → LHb and LHA → LHb projection neurons
Daniel Fürth
Meletis Lab
daniel.furth@ki.se
Content:
- Projection-specific tracing using rabies virus
- n = 3 in each group
- VGluT2-cre:
- 0.7 μl AAV-TVA-V5-G
- 1.5 μl rabies-ΔG-EGFP (3 weeks)
-
GPi → LHb:
- AP: -1.34 mm
- ML: 1.75 mm
- DV: -4.00 mm
-
LHA → LHb:
- AP: -1.00 mm
- ML: 1.00 mm
- DV: -4.50 mm

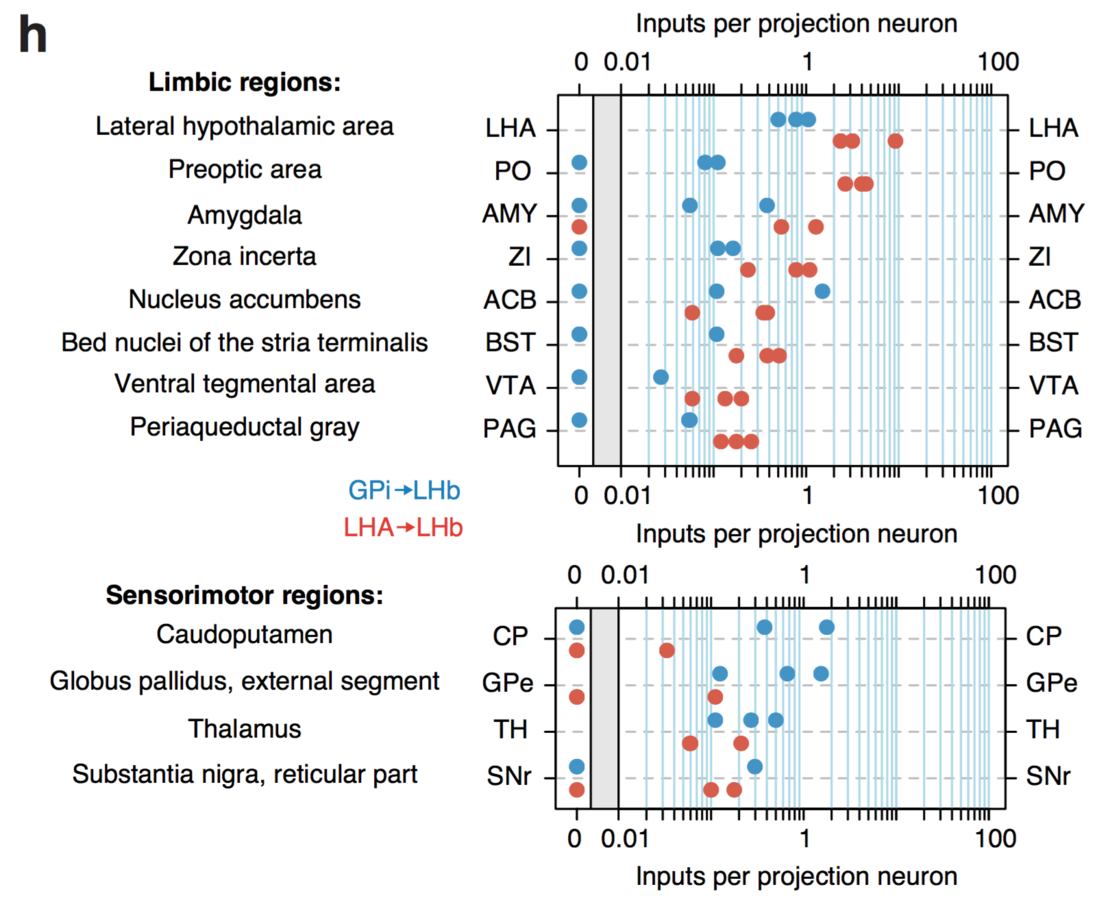


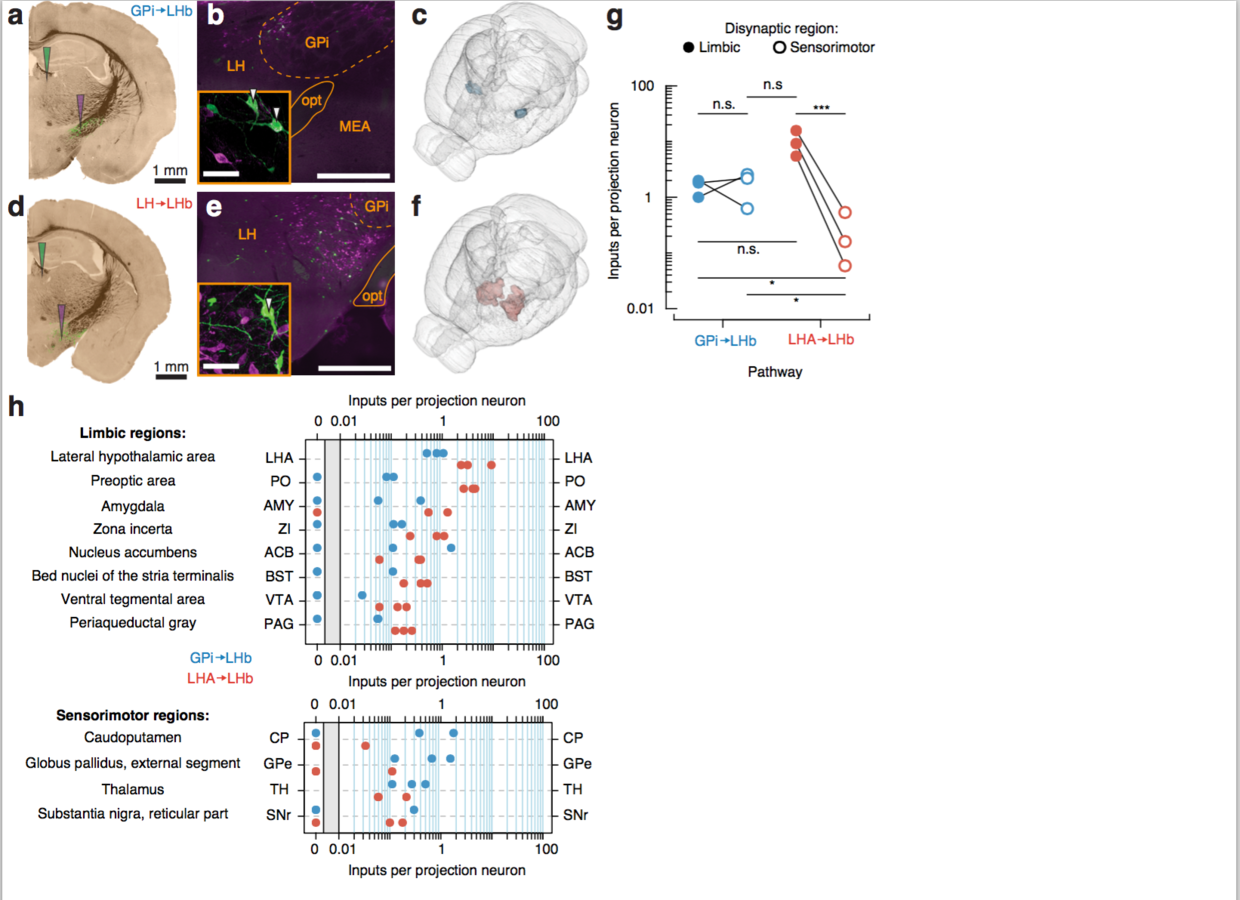
Differential innervation of GPi → LHb and LHA → LHb projection neurons
Daniel Fürth
Meletis Lab
daniel.furth@ki.se
Conclusions:
-
Significant type of region (limbic vs. sensorimotor) times pathway (GPi-LHb vs. LHA-LHb) interaction effect:
-
[F(1,8) = 21.24, P < .01]
-
-
The interaction is driven by reduced input from sensorimotor to the LHA-LHb pathway compared to the GPi-LHb pathway
-
[P < .05]
-
Fit: aov(formula = log10.count ~ GPi.inj * limbic, data = data4)
Df Sum Sq Mean Sq F value Pr(>F)
GPi.inj 1 0.0211 0.0211 0.20 0.66656
limbic 1 2.2651 2.2651 21.50 0.00167 **
GPi.inj:limbic 1 2.2386 2.2386 21.24 0.00173 **
Residuals 8 0.8430 0.1054
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = log10.count ~ GPi.inj * limbic, data = data4)
$GPi.inj
diff lwr upr p adj
GPi-LHA 0.08382212 -0.3483674 0.5160116 0.6665601
$limbic
diff lwr upr p adj
Limbic-Sensorimotor 0.8689223 0.4367328 1.301112 0.0016744
$`GPi.inj:limbic`
diff lwr upr p adj
GPi:Sensorimotor-LHA:Sensorimotor 0.947659617 0.09887367 1.79644557 0.0297026
LHA:Limbic-LHA:Sensorimotor 1.732759802 0.88397385 2.58154575 0.0008203
GPi:Limbic-LHA:Sensorimotor 0.952744428 0.10395848 1.80153038 0.0289255
LHA:Limbic-GPi:Sensorimotor 0.785100186 -0.06368576 1.63388614 0.0701754
GPi:Limbic-GPi:Sensorimotor 0.005084812 -0.84370114 0.85387076 0.9999972
GPi:Limbic-LHA:Limbic -0.780015374 -1.62880132 0.06877058 0.0721037> shapiro.test(data4$log10.count)
Shapiro-Wilk normality test
data: data4$log10.count
W = 0.96947, p-value = 0.9051
> shapiro.test(data4$cell.count)
Shapiro-Wilk normality test
data: data4$cell.count
W = 0.50817, p-value = 2.031e-05
> shapiro.test(data4$cell.count/data4$starter.pop)
Shapiro-Wilk normality test
data: data4$cell.count/data4$starter.pop
W = 0.72223, p-value = 0.001392> bartlett.test(log10.count~interaction(GPi.inj, limbic), data=data4)
Bartlett test of homogeneity of variances
data: log10.count by interaction(GPi.inj, limbic)
Bartlett's K-squared = 2.0333, df = 3, p-value = 0.5655Copy of rania_GPi_LHA_labmeeting_4_July_2016
By Daniel Fürth
Copy of rania_GPi_LHA_labmeeting_4_July_2016
- 661


