To guide the scope of the revisions, the editors discuss the referee reports in detail within the team, including with the chief editor, with a view to identifying key priorities that should be addressed in revision.
In revising your manuscript, please
- clarify the performance of the software in moving between 2D and 3D and "slicing" at arbitrary angles;
- streamline the text to make it more accessible to our broad neuroscience readership (including those without computer science expertise);
- address all other points raised by the reviewers.
Rev #1 Computer scientist:
A technical point that should be clarified is this following. The methodology will achieve its full power using a 3D image block of the mouse brain, as noted, and thenby
referencing locations ....
However, at present atlasing systems such as the Allen Brain Reference Atlas are based on 2D annotations and lofted into 3D. While the Openbrainmap system can cut the 3D brain at arbitrary angles the annotations themselves are not accurate at the same cellular resolution.
Rather the system needs to cut the planes at angles comparable to those used in the Allen Atlas and then to perform a 2D contour outline registration to that section. This workflow issue is central to the single cell workflow and should be clarified for the reader.
Overall a very strong manuscript and important advance in single cell neuroinformatics.
Rev #2 Molecular biologist
From my understanding, this feature enables the “re-slice” of any delineated brain structure in the Allen Reference Atlas.
This is an important feature with the significant potential to improve accuracy of quantifying histological/genetic/viral labeling in different mouse brains (it is difficult to cut all brains consistently in the same angle).
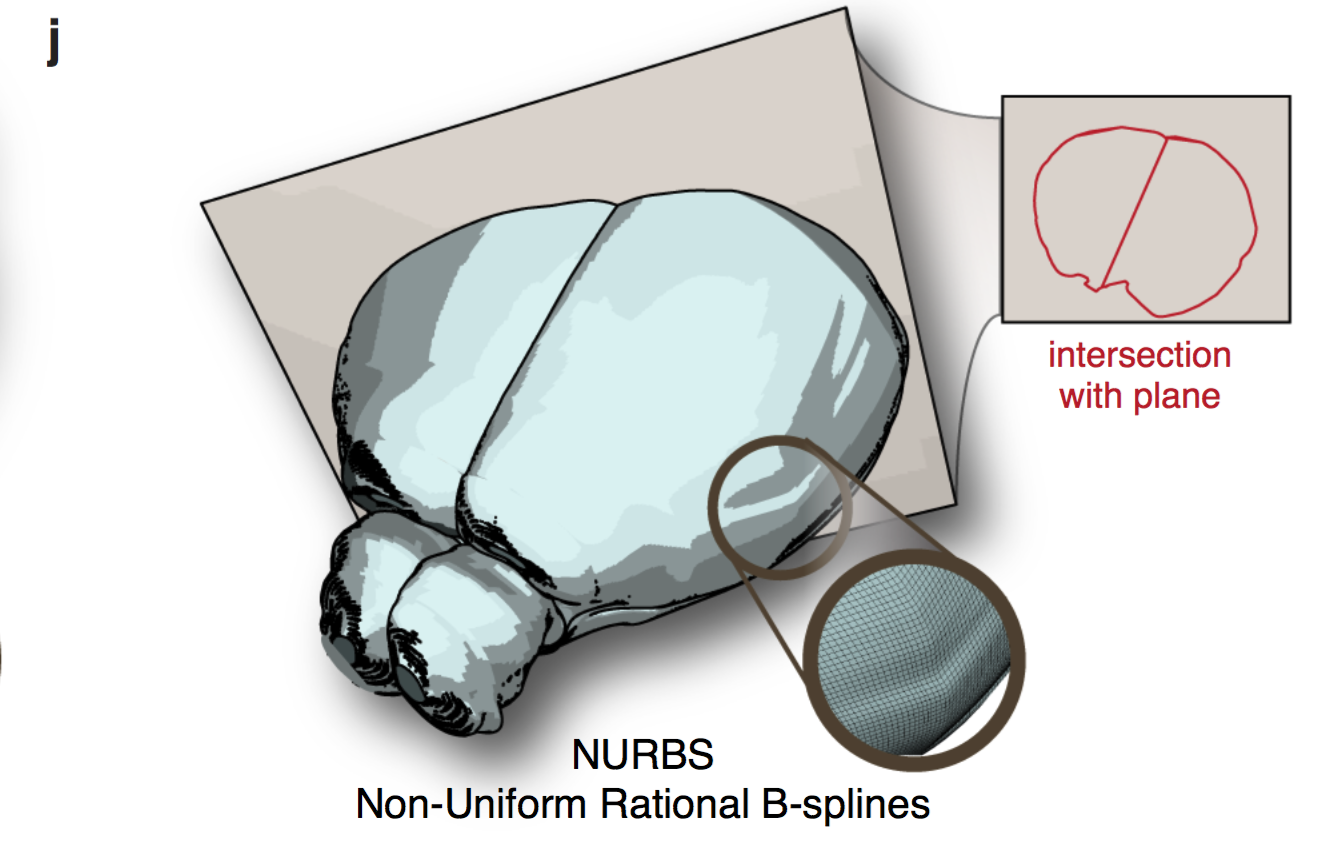
Although the authors demonstrated the principle of the NURBS in Figure 1j, I would like to see what the “re-sliced” atlas looks like in an arbitrarily sliced plane.
There are about 200 µm gap between two adjacent coronal planes in the current version of the Allen Reference Atlas.
How do you ensure the accuracy of delineations and 3D rendering of these structures?
I am impressed by the description of the robustness of the entire software package for mapping and quantifying data of different modality.
Overall, the manuscript is very well written
and all figures are high quality. If successful, this will be a valuable resource for the field, although personally I think this manuscript might be a better fit for Nature Method.
Rev #1 Connectomist
None of these drawbacks are insurmountable.
Yet it seems difficult to understand that the authors would write the paper like that in the first place. It's like they were out of their depth in the understanding of the methods and considered an accomplishment the mere fact that they could put together a working software application.
No comment here that there are many "NIS" systems out there, such as the the brainmaps.org by Shawn Mikula, the neurodata.IO by Vogelstein at Hopkins University (formerly open connectome project), the Allen Brain Atlas software, the Virtual Fly Brain by the Armstrong group in Edinburgh, the Fruit Fly Brain Observatory by the Lazar group at Columbia University in New york, the CATMAID software for synaptic-level neural circuit reconstruction and analysis, the Whole Brain Catalog by Larsson, and the http://opensourcebrain.org/, in addition to the worm atlas by David Hall and colleagues.
deck
By Daniel Fürth
deck
- 723


