Boosting one tree
at a time
Ferran Muiños
updated: Monday 20210301

Aim of the talk
"Demystify tree ensemble methods and in particular boosted trees"
-
Quick overview of regression
-
Gentle intro to decision trees and tree ensembles
-
What does boosting intend?
-
Some examples along the way
Why ensemble methods in the end of the day?
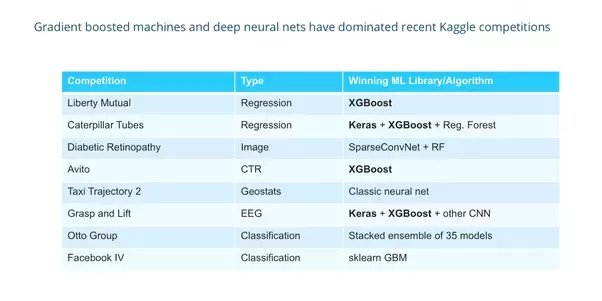
https://www.quora.com/What-machine-learning-approaches-have-won-most-Kaggle-competitions
Regression
covariates
response


Regression
covariates
response
Problem statement:
Find a function that gives a precise description of the dependence relationship between and :
Regression
Alternative problem statement:
Given a collection of samples
find a function that provides:
- Low-error approximations (good fit)
- Expected good fit for any dataset of the same kind.
covariates
response
First example: cell culture

First example: cell culture

What patterns do we see?
First example: cell culture


First example: cell culture
Average Smoothing
Nadaraya-Watson

: bell-shaped kernel


First example: cell culture
Linear
Quadratic
First example: cell culture
Parametric methods:
- assume global shape
- very restricted overall
- maybe inaccurate prediction
- maybe easier to interpret
Smoothers:
- agnostic shape
- shape is locally restricted
- useful for prediction
- more difficult to interpret


Whatever the strategy, we want some
that satisfies
Whatever the strategy, we want some
that satisfies
Applicable to datasets with n covatiates
Whatever the strategy, we want some
that satisfies
This is the key!
How do we know what to expect after all?
Data subsets
Dataset
Training dataset
Test dataset
Fit the model
Test the model




Bagging
Bagging = Bootstrap + Aggregating
- Pick several random subsets of samples:
- Train a model with each subset:
- Create a consensus model:
"Averaging" is the typical way to reach consensus

Bagging does a decent work even with weak components
Me learn good
Tree Ensembles
Trees functions
a.k.a. decision trees:
- Have a root where the input goes
- Leaves contain values
- Inner nodes are if-else statements
- If-else conditions are of the form
Example
yes
no
yes
no

1st split
2nd split
What is the best least-squares fitting stump?

?
- Root splits the data:
- Set leaf values:
?
Which split gives minimum loss?
e.g. loss = RSS
What is the best least-squares fitting tree?
?
- Root splits the data:
- Set leaf values:
For which split do we get minimum RSS?

Bagging with stumps...






Random Forests

from sklearn.ensemble import RandomForestRegressor model = RandomForestRegressor(n_estimators=3, max_depth=1) res = model.fit(temp, rate)
Random Forests
n_estimators=1000, max_depth=1


n_estimators=1000, max_depth=2
Gradient Boosting
- Ensemble model
- General framework where weak learners can take any form
- Taking trees as weak learners gives a greedy version of Random Forest
- Derivative of the loss function plays a prominent role --whence the "gradient".
How it works (XGBoost)
- Training Samples:
- Set a loss function e.g.
Instead of fitting a global model to the loss function, training is done by adding one tree at a time (additive training):
- Initialize the model with the constant tree
- Sequential growth. At each step add a new tree
How it works
Goal: find that minimizes the loss
The regularization term penalizes the tree complexity.
For example, in XGBoost:
is the number of leaves
are the values (or weights) at the leaves
?
How it works
We can provide a second order
Taylor approximation of the Loss function.
- Recall:
- Define:
How do we find ?
?
How it works
How do we find ?
New goal: minimize this new loss function
Regrouping by leaf, we can write it as a sum of quadratic functions, one for each leaf:
?
How it works
How do we find ?
If the tree structure of t is fixed, then the optimal weights for each leaf are given by:
where and
Evaluating the new loss on gives a scoring on each possible tree structure.
Trees are grown greedily so that this scoring keeps decreasing at every step.
?
How it works
Example of a tree structure:
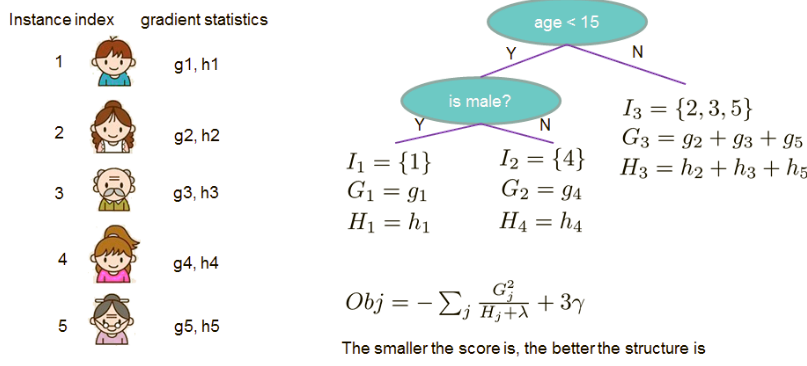
How it works
Now that we have a way to measure how good a tree is, ideally we would enumerate all possible trees and pick the best one. In practice, however, this is intractable.
?
?
?
Is the new split beneficial or not?
Optimal tree structure is searched for iteratively
Tunning hyperparams
- loss function
- learning rate a.k.a. shrinkage:
- number of estimators (trees)
- maximum depth of trees
- randomization rules:
- subset of samples (bagging, out-of-bag error)
- per-tree/per-split subset of covariates
- regularization parameters
We can introduce rules to constraint the search for each update. These rules define the "learning style" of the model.
Why Tree Ensembles?
Upsides:
- Non-parametric (shape agnostic)
- Up to a variety of regression and classification tasks
- Modelling flexibility
- Admit a large number of covariates
- Good prediction accuracy
- Good at capturing interactions between covariates by design
- Interpretation is feasible: ranking variables, partial dependence
- Efficient functions
Downsides:
- Steeper learning curve for users
Why gradient boosting?
Upsides:
- Same reasons why I like Random Forests, plus...
- Very good accuracy with fewer learners (greedy).
- Excellent XGBoost implementation (R, Python).
- Many model design options at reach.
Downsides:
- Sequential by design, hence intrinsically slower to train than other methods like RF.
- Hyperparameter tuning.
References
-
Freund Y, Schapire R A short introduction to boosting
-
Hastie T, Tibshirani R, Friedman J The Elements of Statistical Learning
-
Natekin A, Knoll A Gradient boosting machines, a tutorial
-
XGBoost Documentation: https://xgboost.readthedocs.io/en/latest
Gradient boosting seminar at Bioinfo UPF
By Ferran Muiños
Gradient boosting seminar at Bioinfo UPF
A gentle introduction to ensemble tree models and gradient boosting for regression.
- 1,120



