Secondary structure prediction using neural networks
Secondary structure prediction
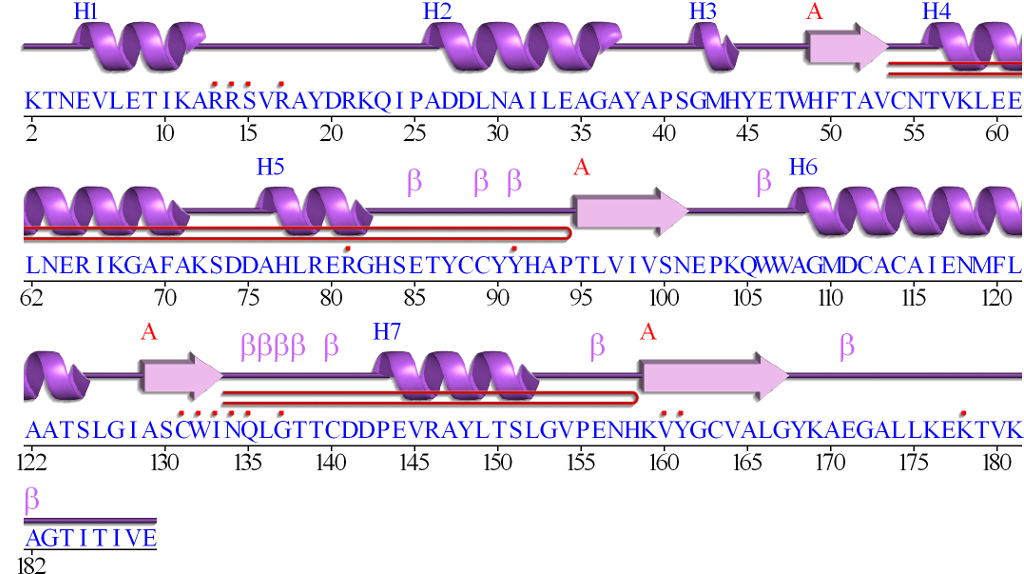
Secondary structure prediction as a sequence-to-sequence translation
NQGKIWTVVNPAIGIPALLGSVTVIAILVHLAILSHTTWFPAYWQGGV
Amino acids:
CTTTGGGCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHCCC
Sec. structure:
There are 20 amino acids, and 8 secondary structures.
These are the letters in the sequences
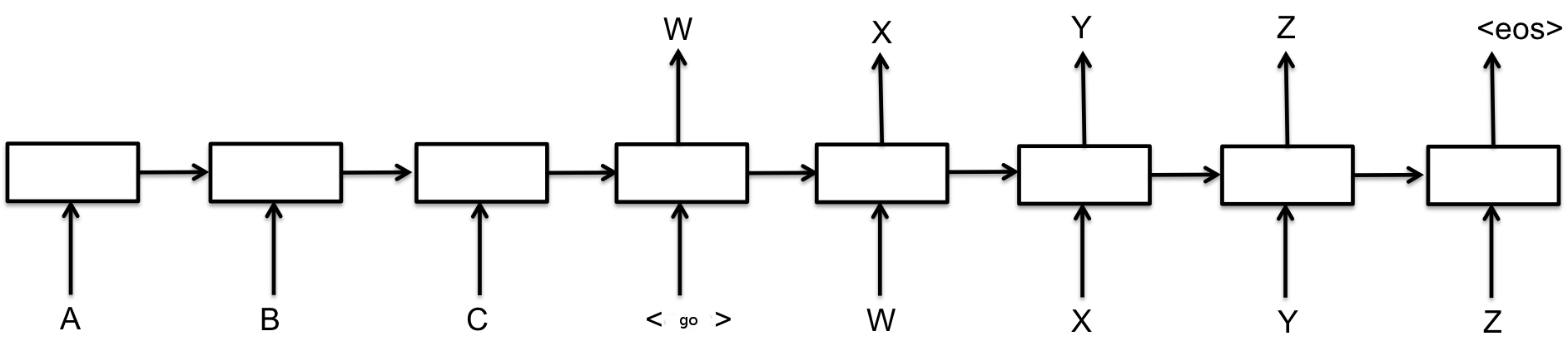
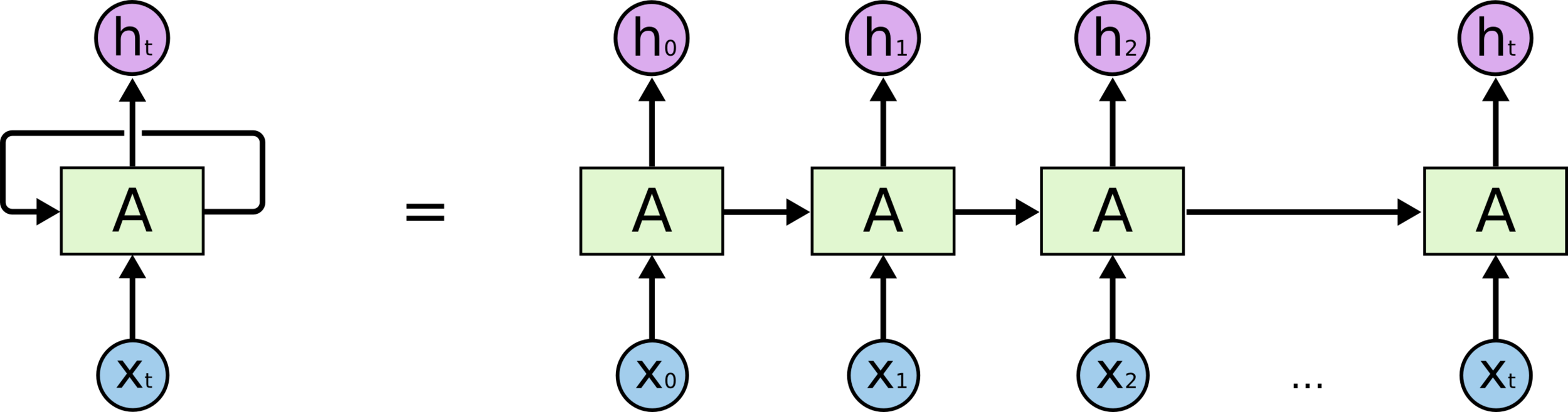
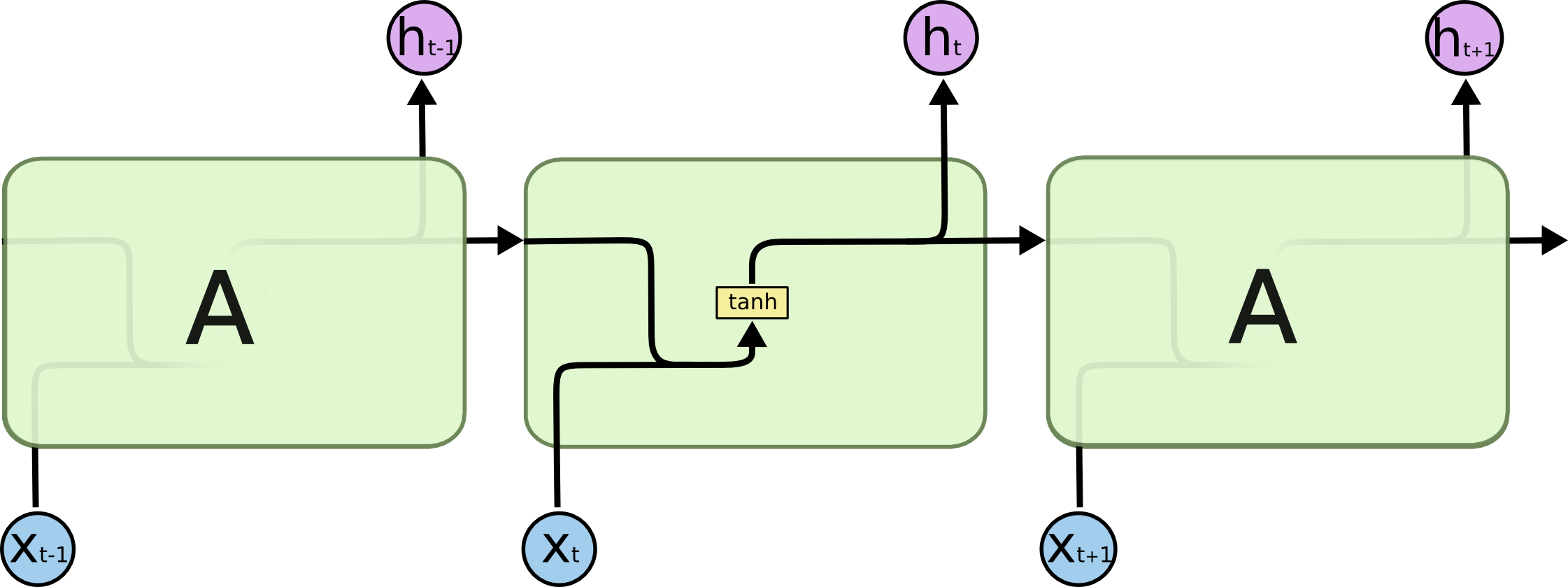
Recurrent neural networks
Sequence-to-sequence models
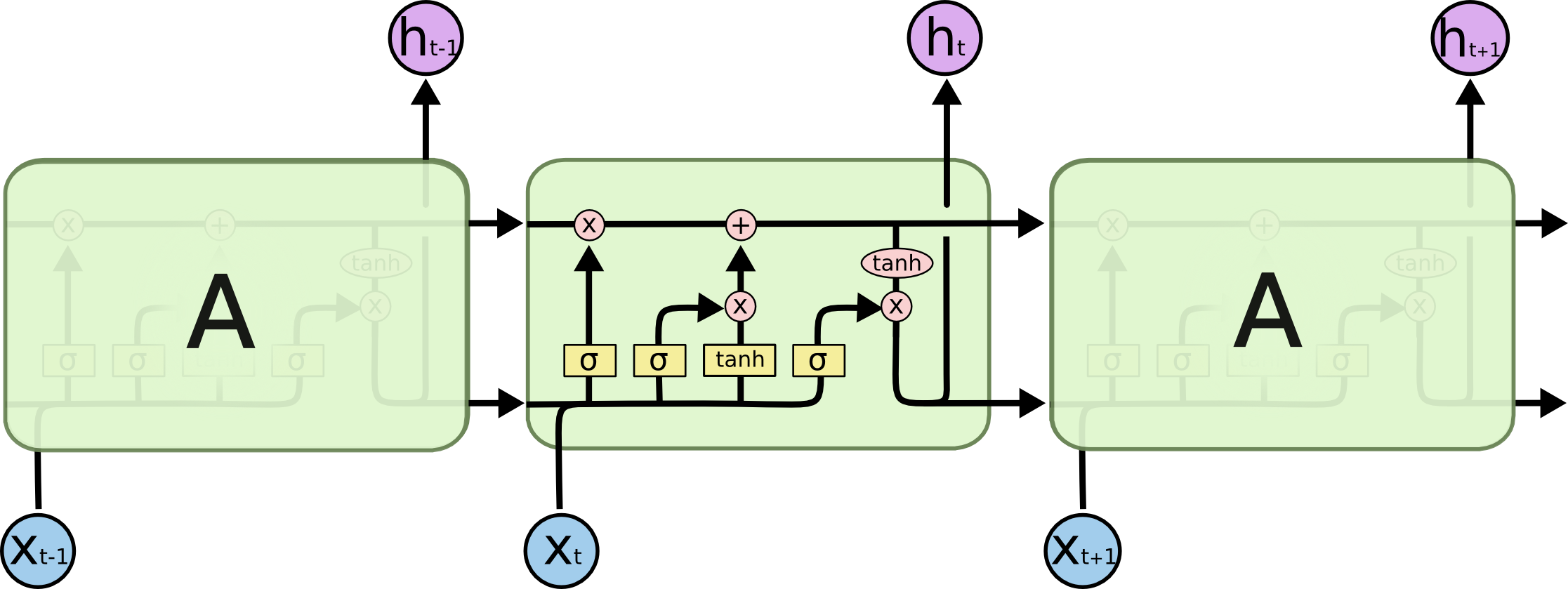
Long short-term memory
Long short-term memory
Long short-term memory

TensorFlow
Preparing the data



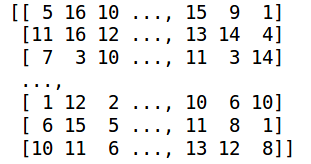

Input (trainX)
Output (trainX)
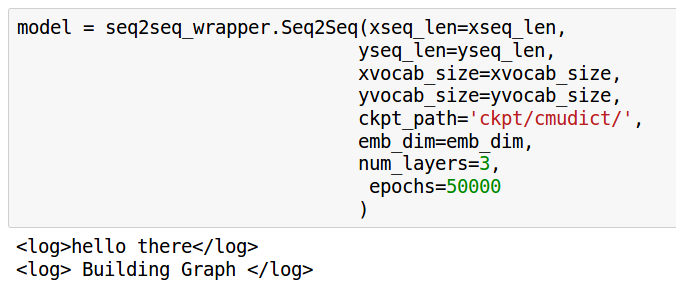
Building the model
a.k.a. computational graph
Training
Things that produce minibatches
Tell tensorflow to train!
~~Complicated backprop under the hood~~
Testing

= fraction of all characters that it has gotten wrong
Results
81.3% accuracy
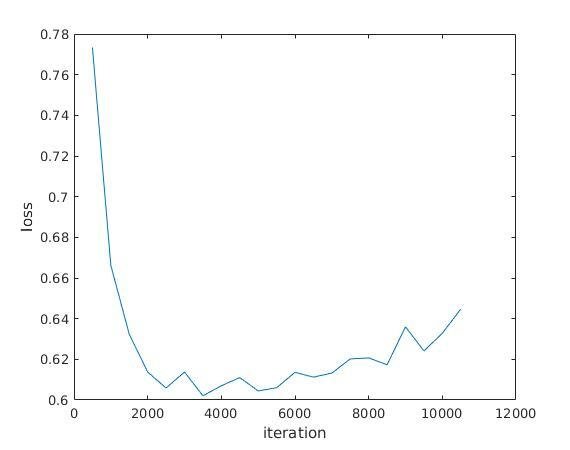
Length 60 segments
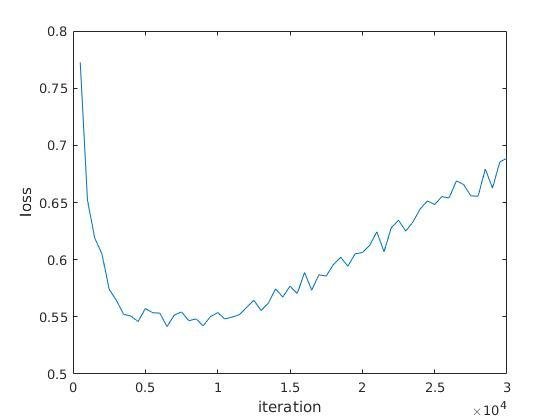
Length 100 segments
61% accuracy
Next steps
- CB513/CB6133 datasets
- Inconsistent sequence length handling
- GPU
ssnn
By Guillermo Valle
ssnn
- 510



