What you should know about
Graph-based Reference Genomes
What are they?
Why should they be used?
How can you use them?
What are they?
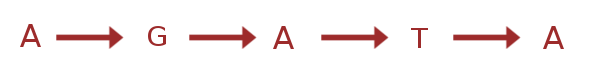

(linear) reference genome
Also common in the population
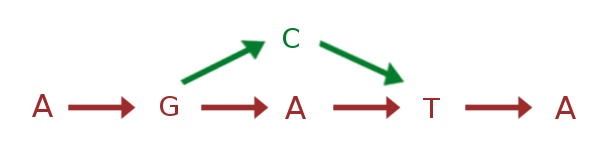
Graph based reference genome containing both sequences
.
What are they?
UC Santa Cruz Genomics Institute
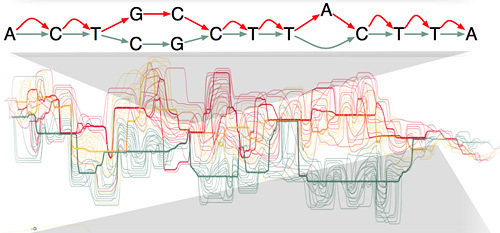
Chr17, GRCh38
Alt loci
Alt loci
GRCh38 can be seen as a graph
Why use them?
- Sequence differs between individuals
- Enables more accurate mapping and variant calling
- Create reference for variation rich species
To account for natural genetic variation, we consider the more general case in which a reference genome is represented by a graph rather than a set of phased chromosomes; the latter is treated as a special case.
Paten et al. (2014)
Some examples
Reference genome of malaria-infected mosquitoes

Harding et al.
- Genomes from over 700 mosquitoes
- Used for variant calling
De novo assembly and genotyping of variants using colored de Bruijn graphs
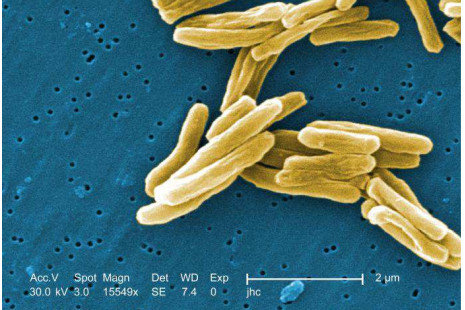
Population reference graphs for tuberculosis bacteria
Mouzos et al.
- 300 tuberculosis genomes in one graph
- Used for mapping and visualization
Iqbal et al., Nature Genetics 2012
- Variant calling on a human graph-based reference genome
How can you use graph-based reference genomes?
You can use GRCh38, however:
- Few tools use the alternative loci
- Where to find annotated data?
You can create your own using vg:
- Building a graph with 1000 genomes vcf requires 200+ GB of memory and ~1.5 TB of disk space
- Compiling vg can be difficult
- No annotated data

What we have done
Summary
- Graph-based reference genomes improve mapping, variant calling and further analysis on genomic features
- Using GRCh38 and its alternative loci is a good start
- You can map to GRCh38 using BWA-MEM, Hisat2 or vg
- Using a linear reference increases noise or even leads to biases when doing statistical genomics
- vg: github.com/vgteam/vg
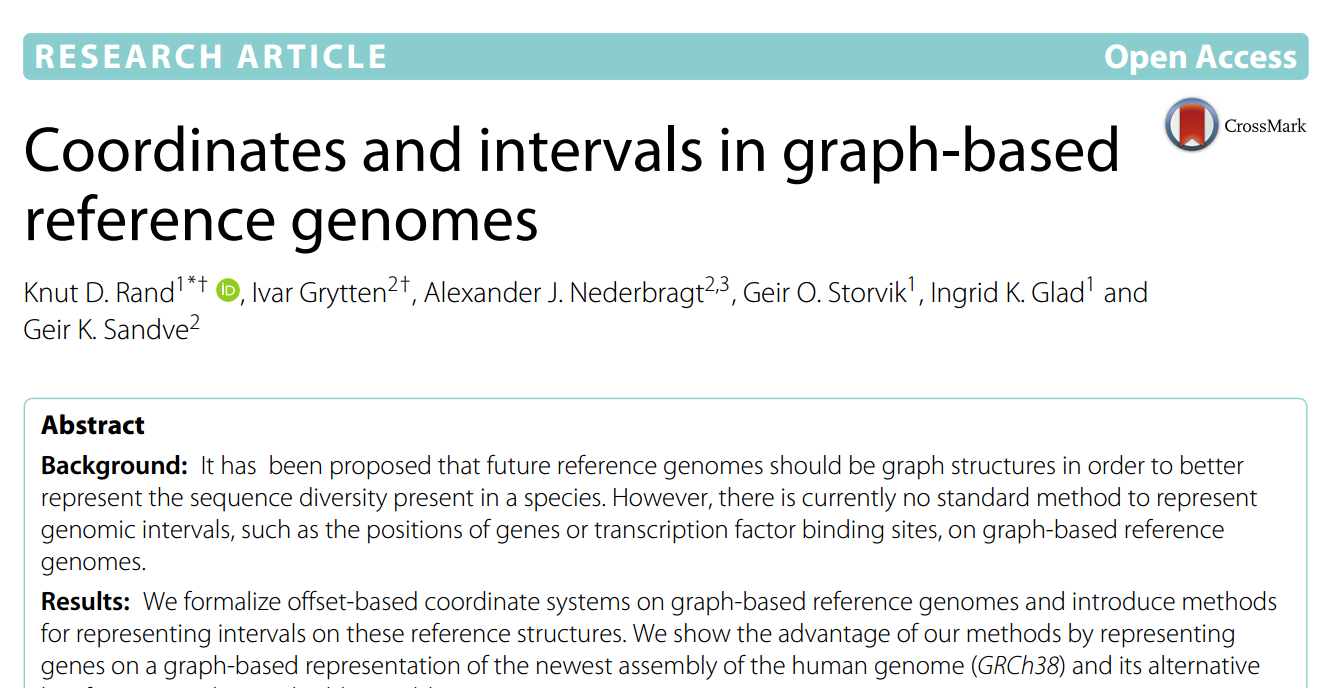
- Our article: Coordinates and intervals in graph-based reference genomes
- Guide on how to map to GRCh38:
http://gatkforums.broadinstitute.org/gatk/discussion/8017/how-to-map-reads-to-a-reference-with-alternate-contigs-like-grch38
Check out:
grbgs_short
By ivarg
grbgs_short
- 463