HDF5 in Python
Giacomo Debidda
18/12/2017 @PyData Munich
Topics
- HDF5
- HDF5 tools
- h5py
- PyTables

- Data Model
- Library
- Format
HDF5 is...
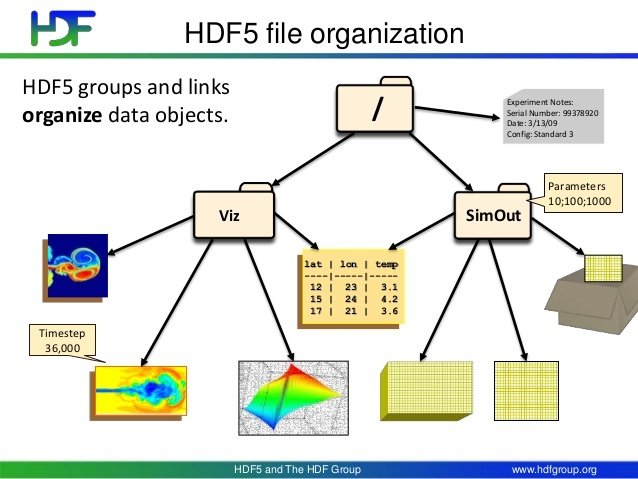
A filesystem in a file
/ root group (every HDF5 file has a root group)
/foo member of the root group called foo
/foo/bar member of the group foo called bar
Working with groups and group members is similar to working with directories and files in UNIX.
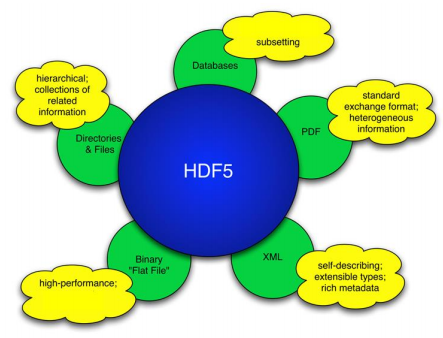
HDF5 Data Model
- Datasets (i.e. files in a filesystem)
- Groups (i.e. directories in a filesystem)
- Attributes (i.e. metadata of file/directory)
The HDF5 File Format Specification specifies the bit-level organization of an HDF5 file on storage media.

Why use HDF5?
- Portable
- Self-describing
-
Can contain binary data (in many representations)
-
Allows direct access to parts of the file without first parsing the entire contents
- Supports large/complex/heterogeneous data
- File format tool kit (you can design your own file format and use HDF5 under the hood)
Who uses HDF5?









HDF5 Tools
- h5ls
- h5dump
- h5stat
- h5diff
- HDFView
- HDF Compass
- ViTables
Reference
h5py
-
Thin, pythonic wrapper around HDF5
-
HDF5 errors are converted into Python exceptions
-
Written in Cython
-
Uses numpy objects



Higher level abstraction
It does not want to be a complete wrapper for the entire HDF5 C API
Can be faster than h5py, thanks to out-of-core querying
Allow indexing and complex queries
Built-in compression
Undo mode
Scipy 2015
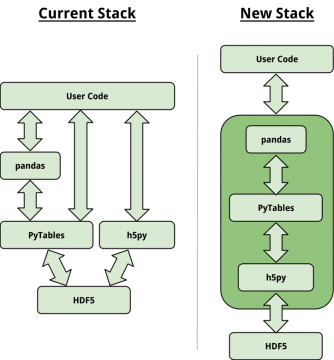
At SciPy 2015, developers from PyTables, h5py, the HDF Group and pandas decided to start a refactor: PyTables will depend on h5py for its bindings to HDF5.
Code, plz!

HDF5 talk PyData
By Giacomo Debidda
HDF5 talk PyData
- 1,600



