Full-Community Metabarcoding: An Ecosystem Map
CBIOMES Annual Meeting 2022
Jesse McNichol (he/him) : PhD, Biological Oceanography
Postdoctoral Scholar, University of Southern California




- Comprehensive (primers target all cellular life)
- Sensitive with deep sequencing
- Specific with "denoising" algorithms




Microbe art: @claudia_traboni
Full-Community Metabarcoding: The Promise
Full-Community Metabarcoding: The Challenges
?
✓
✓
✓
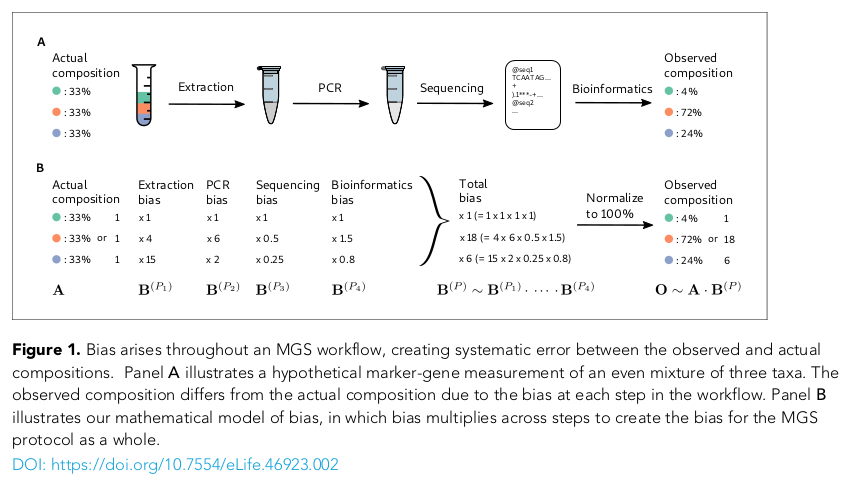
- Biases exist, but are consistent & correctable
- Extraction bias for complex marine mixtures unknown, needs validation (BioGEOSCAPES)
1. Led in silico method optimization
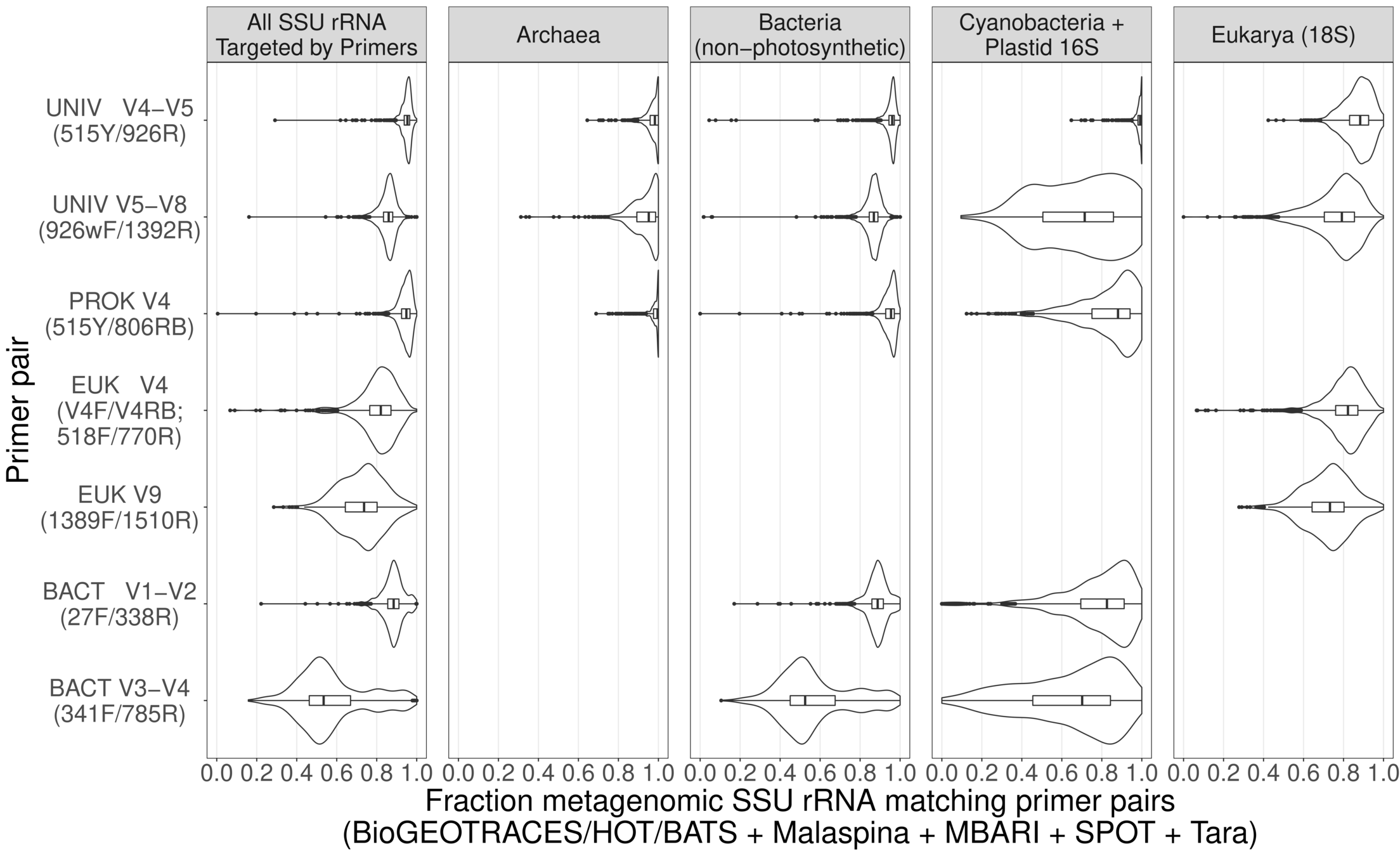
Universal primers for barcoding work almost* perfectly across global oceans
2. Developed collaborations and generated data




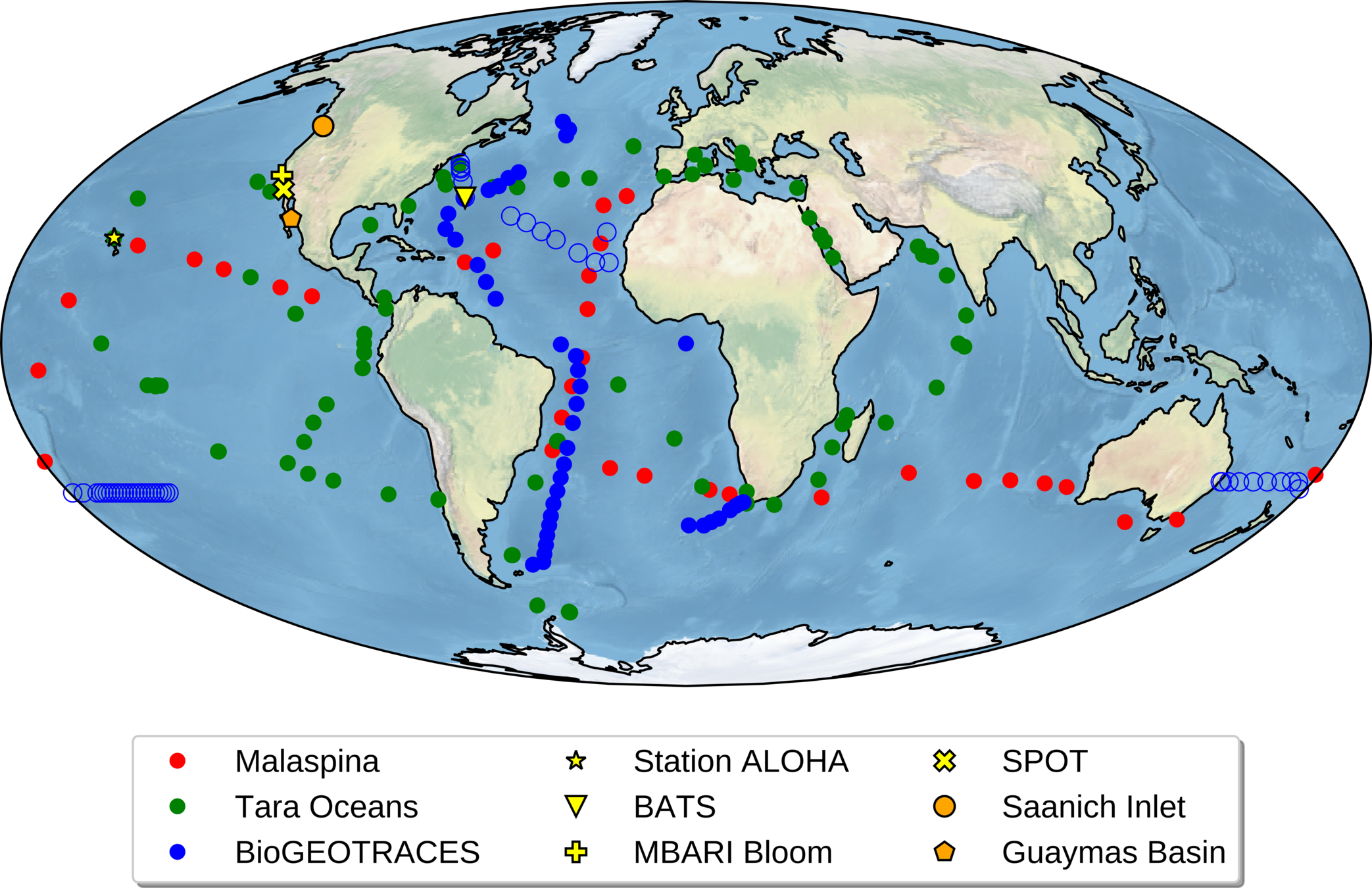
Global metagenomes
Over 800 globally-distributed barcode samples allow model-data intercomparison
Craig Carlson, UCSB
Full-Community Metabarcoding: Progress

Microbe art: @claudia_traboni
p16S
e16S
18S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
Full-Community Metabarcoding: Applications

Microbe art: @claudia_traboni
18S
p16S
e16S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
Full-Community Metabarcoding: Applications
Craig Carlson, Elisa Halewood, UCSB
Fraction of 18S amplicon sequences

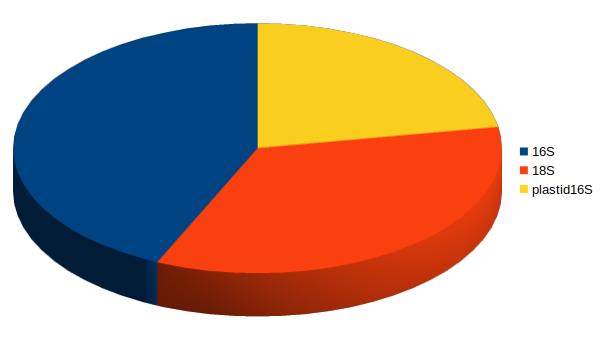
16S
plastid 16S
18S




Jan-Feb 2005
Feb-Mar 2006
With deep sequencing, good coverage for all 3 domains
Full-Community Metabarcoding: Applications



Taxa-specific activity
Full-Community Metabarcoding: An Ecosystem Map






"eDNA"
Full-Community Metabarcoding: Synergies
Research questions
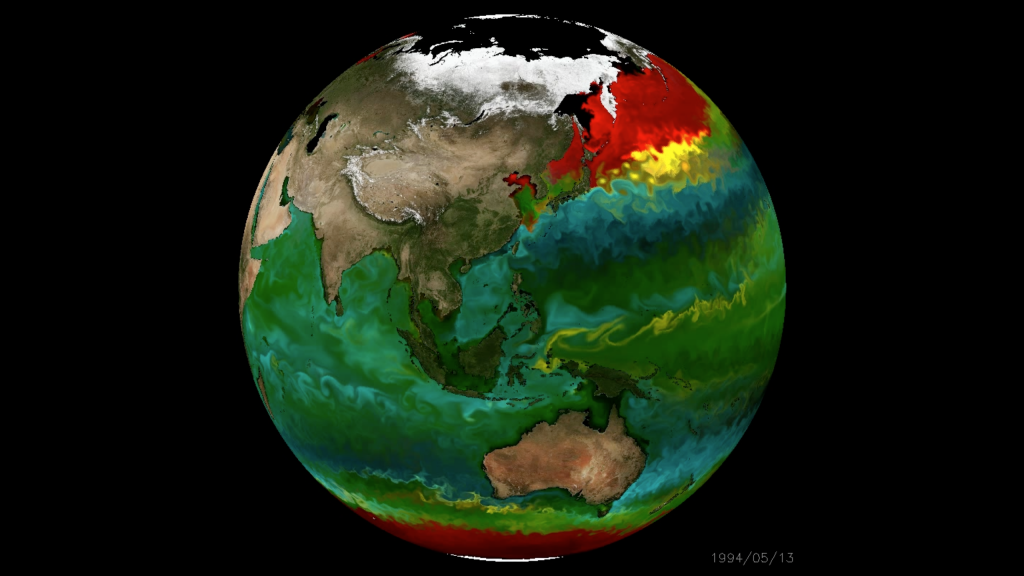
An in silico global phytoplankton model (DARWIN project, MIT)
- How robust are model predictions?
- What is the function of microbial "black box"?
- Can models and data help predict consequences of climate change?
- How well do imaging-based methods (flow cytometry, imaging flow cytobot) align with 3-domain metabarcoding?
Advantages:
- Simple but impactful
- Simons CMAP database makes possible
- Extend by mining global distribution data


Kalmbach et al. (2017), arXiv:1703.07309v1

Example research project #1
Example research project #2
- How can we infer changes in microbial community composition change across decadal time-scales using barcoding data from different regions of SSU rRNA?
Advantages:
- "Rosetta stone" allows intercomparisons, FISH probe design from barcodes
- Address previously impossible questions
- "Data science" experience for student/postdoc

Dueholm et al. (2020) mBio, e01557-20
Database of full-length 16S rRNA


- Taxonomy (CARD/HCR-FISH) / transcription (FISH-TAMB)
- Respiration, cell viability (RSG); Polyphosphate content (DAPI)
Automatic cell-sorting + environmental databases = taxon-specific activity or genetic potential measurements
Many uses for sorted cells:
- Taxon-specific isotope uptake
- Mini-metagenomes with few cells
- Cell isolation for cultivation
Pjevac et al (2019) 10.1111/1462-2920.14739
Multiple sorting axes now possible

Microscale cell sorting enabled by new databases
Applications "at sea"
-
"Rosetta stone" => FISH probes:
- Cell enrichment for mini-metagenomes
- Visualize predicted symbioses / other ecological interactions
- Measure cell-specific traits such as biovolume
-
Attempt cultivation of "microbial dark matter":
- Flow cytometry with "live stains"
- Enrichment experiments
- Intercalibration, intercomparison
- Methods development
- Interdisciplinary by nature
- Lots of room for young scientists to make major contributions


BioGeoSCAPES

Motivated by goal of understanding the Earth System

We anticipate that as trait-based biogeography continues to evolve, micro- and macroorganisms will be studied in concert, establishing a science that is informed by and relevant to all domains of life.” -Green, Bohannan, and Whitaker, Science (2008)
Ecosystem-centered research


Sebastián & Gasol (2019), 10.1098/rstb.2019.0083 ; McNichol et al (2018) 10.1073/pnas.1804351115 ; Zehr (2015) 10.1126/science.aac9752; Bramucci et al (2021) 10.1038/s43705-021-00079-z

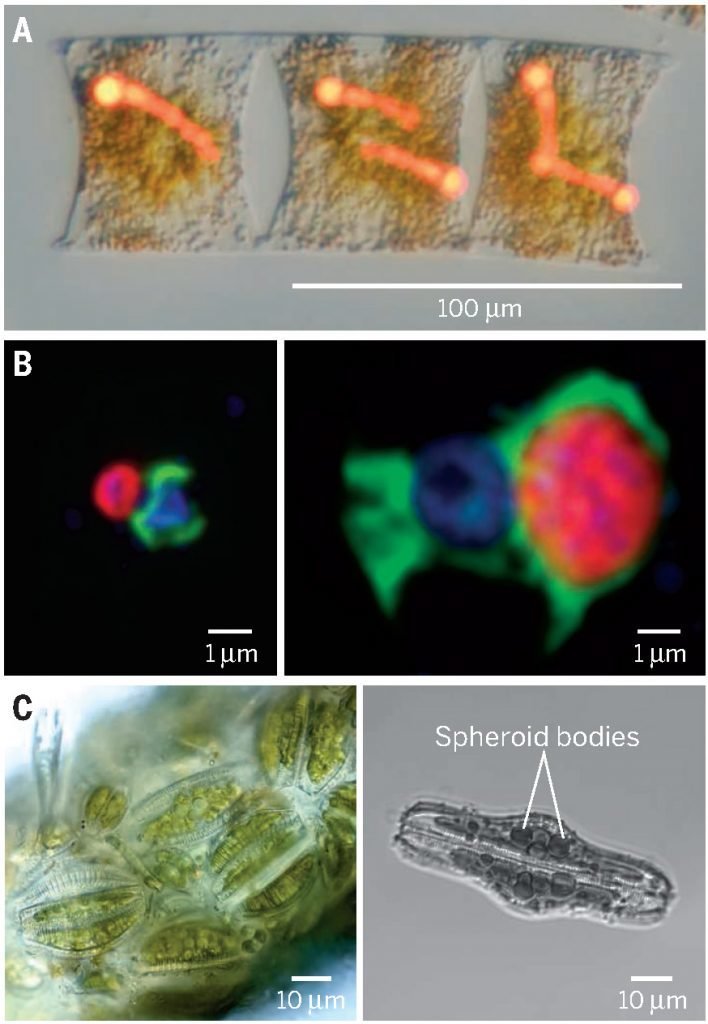

Techniques for microscale ecology & biogeochemistry
Model-data intercomparison (Yubin Raut, USC)



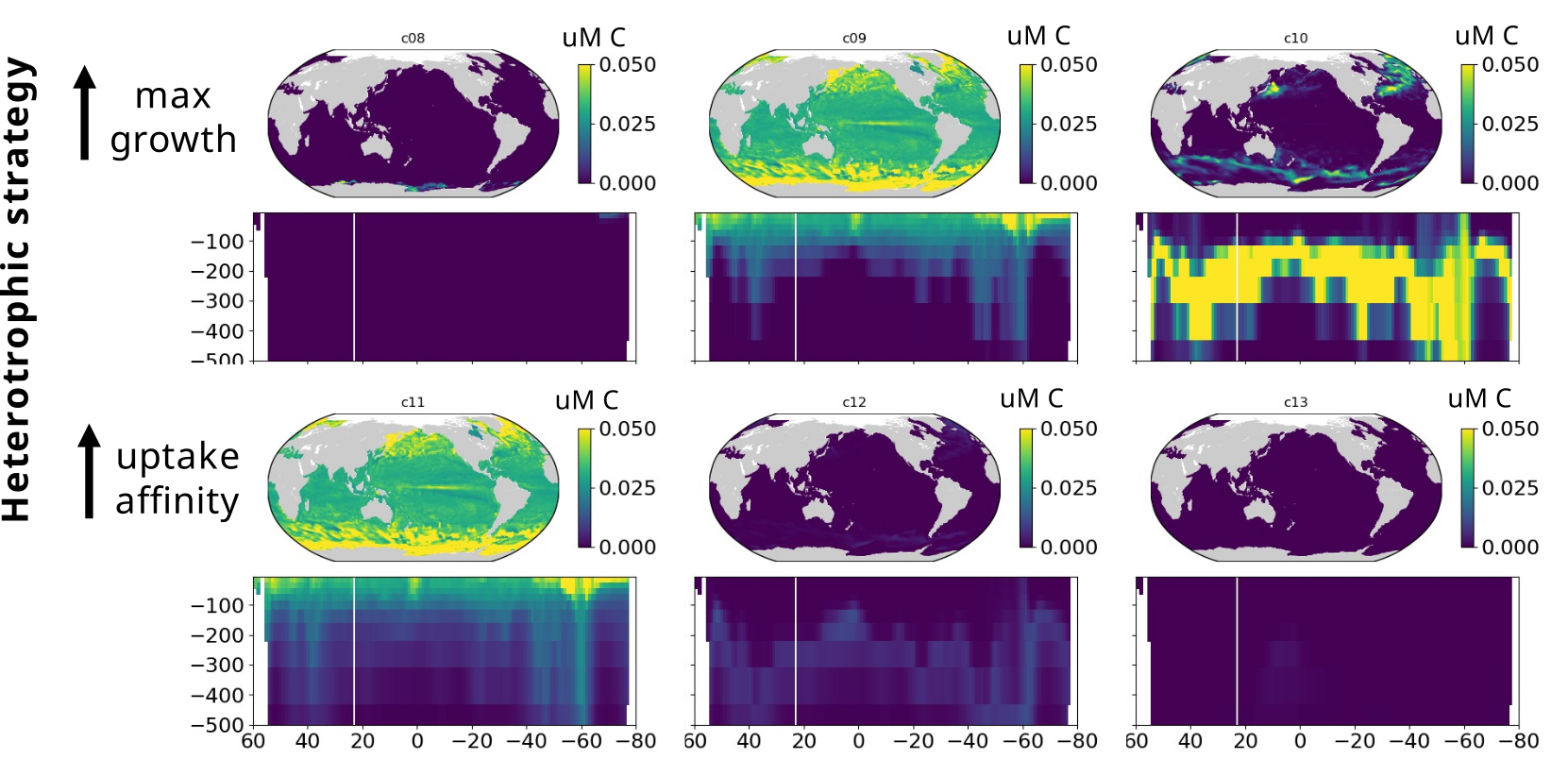
Modelling marine heterotrophs (Emily Zakem, Carnegie Inst.)
Collaborations

Looking for interactions
plastid
16S
mito 16S
nuclear 18S
Space / time
Abundance
A eukaryotic phytoplankter

Models
Microbe art: @claudia_traboni
Ecosystem
Kalmbach et al. (2017), arXiv:1703.07309v1



Physics
Biology
Chemistry

Long term: holistic vision




Jan-Feb 2005
Feb-Mar 2006
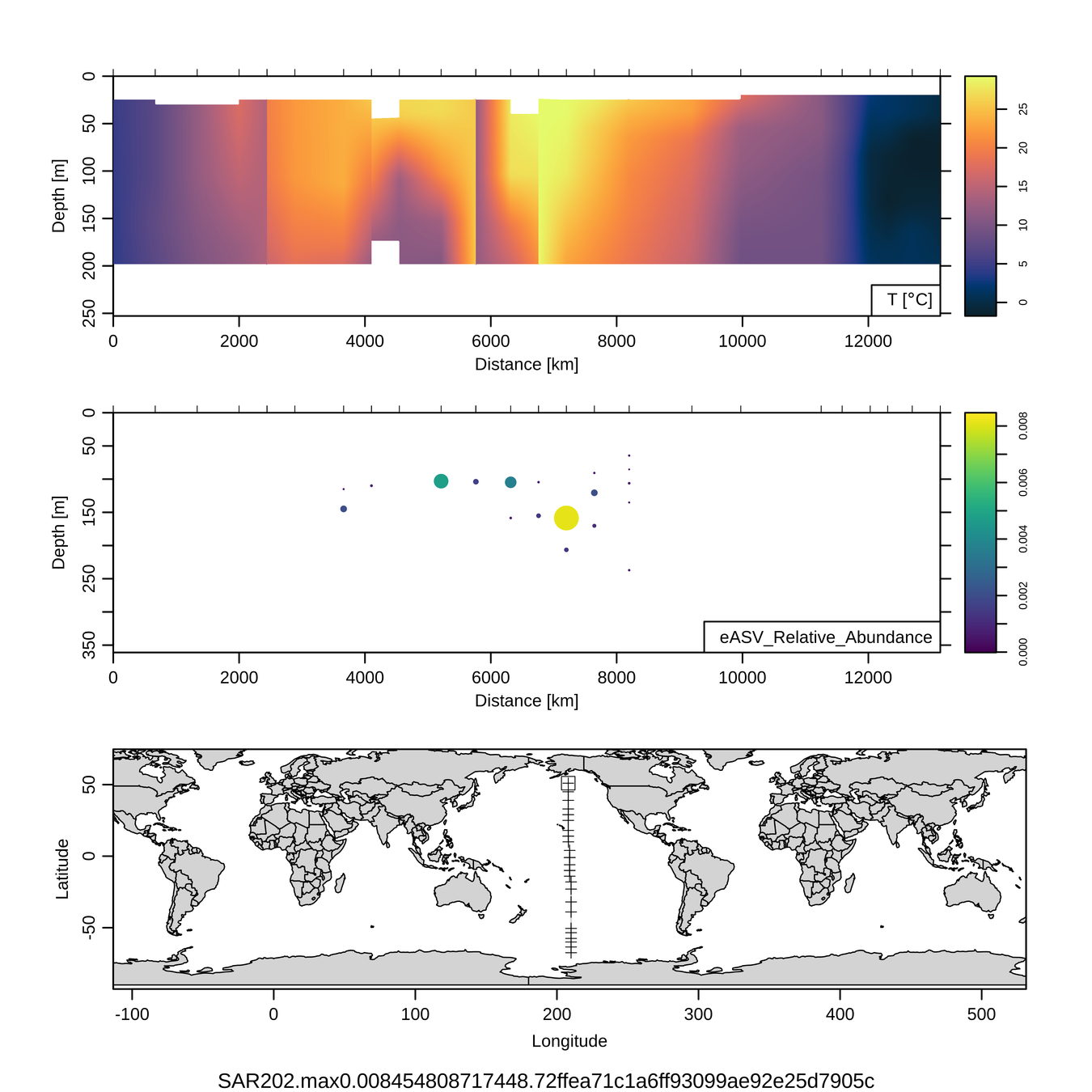
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
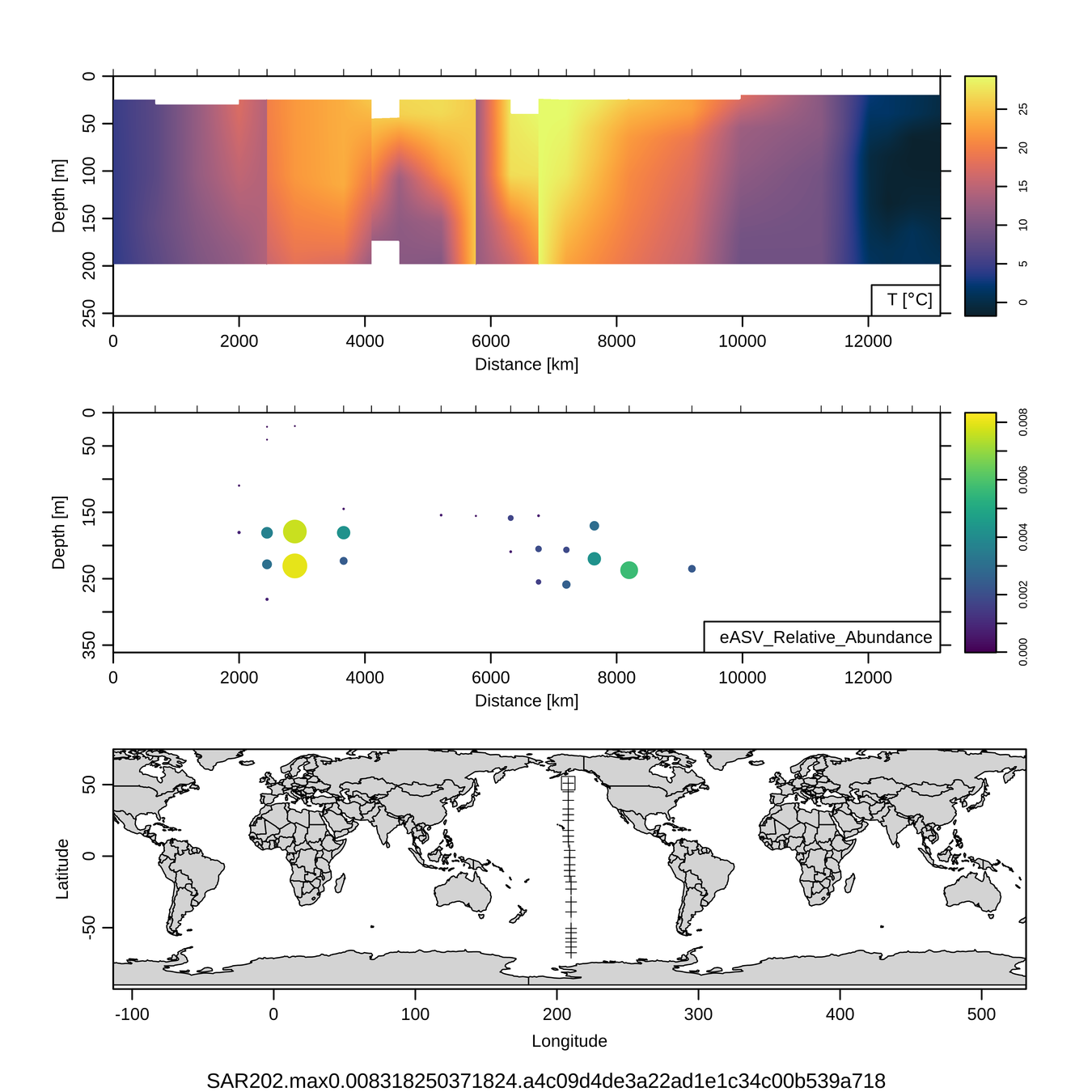
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
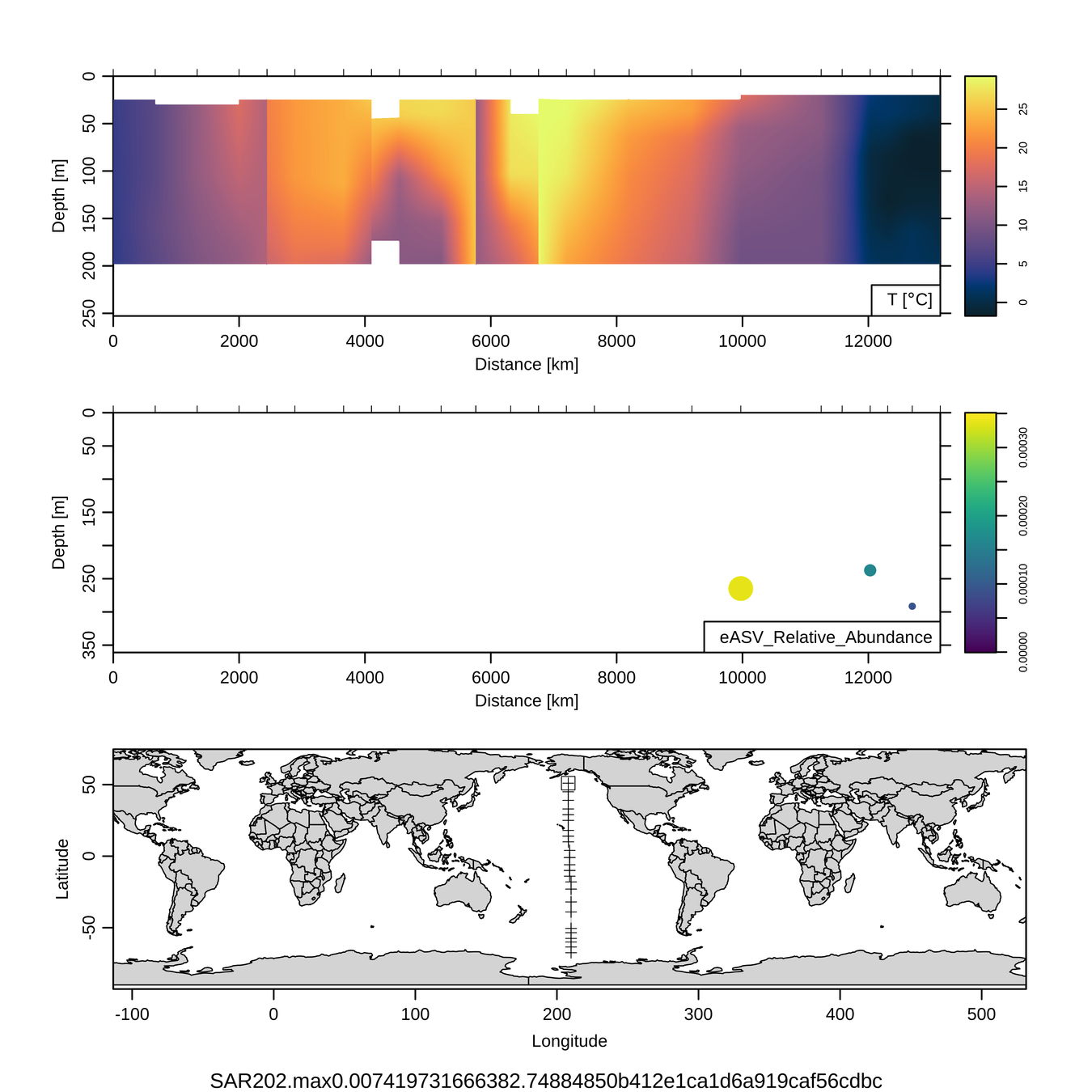
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
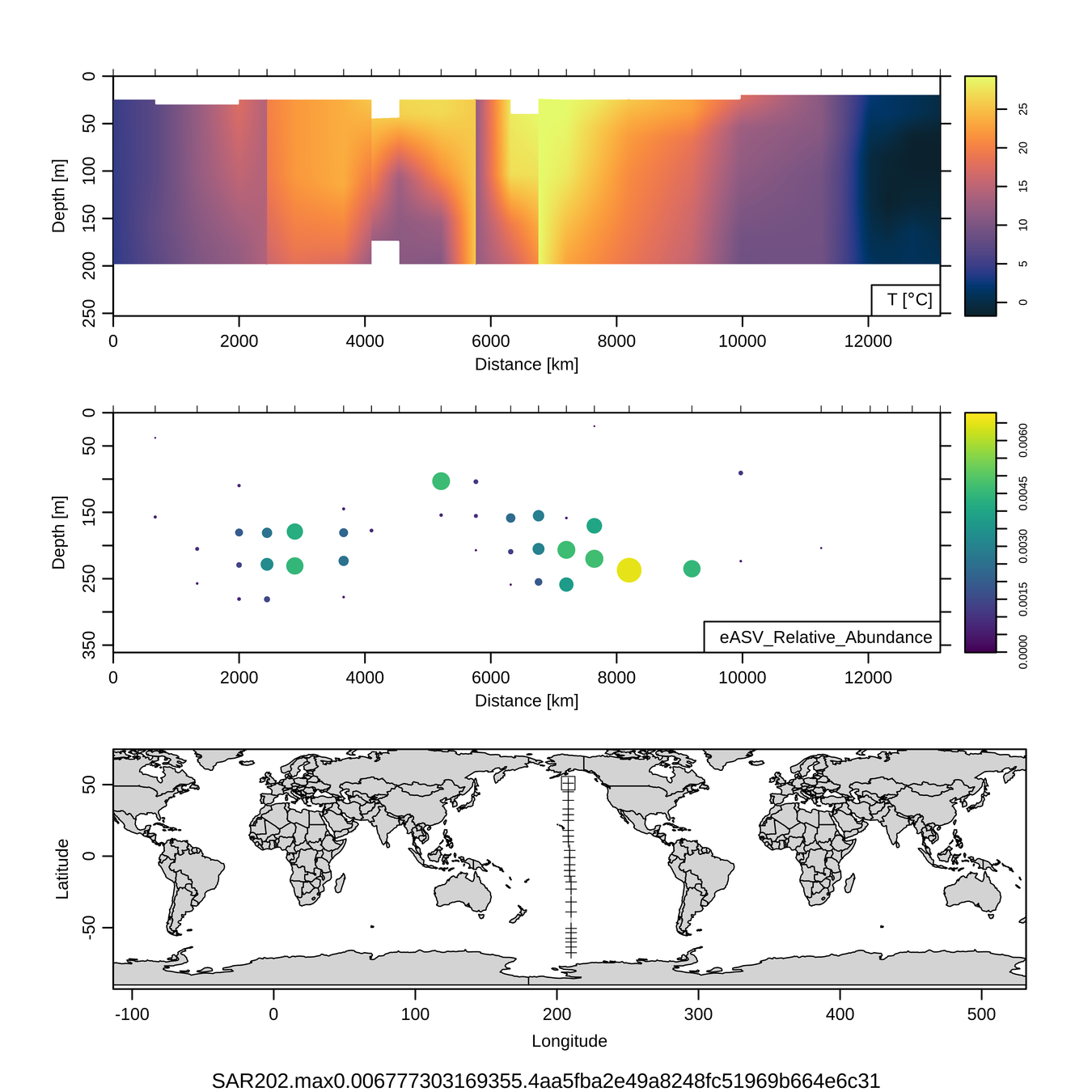
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
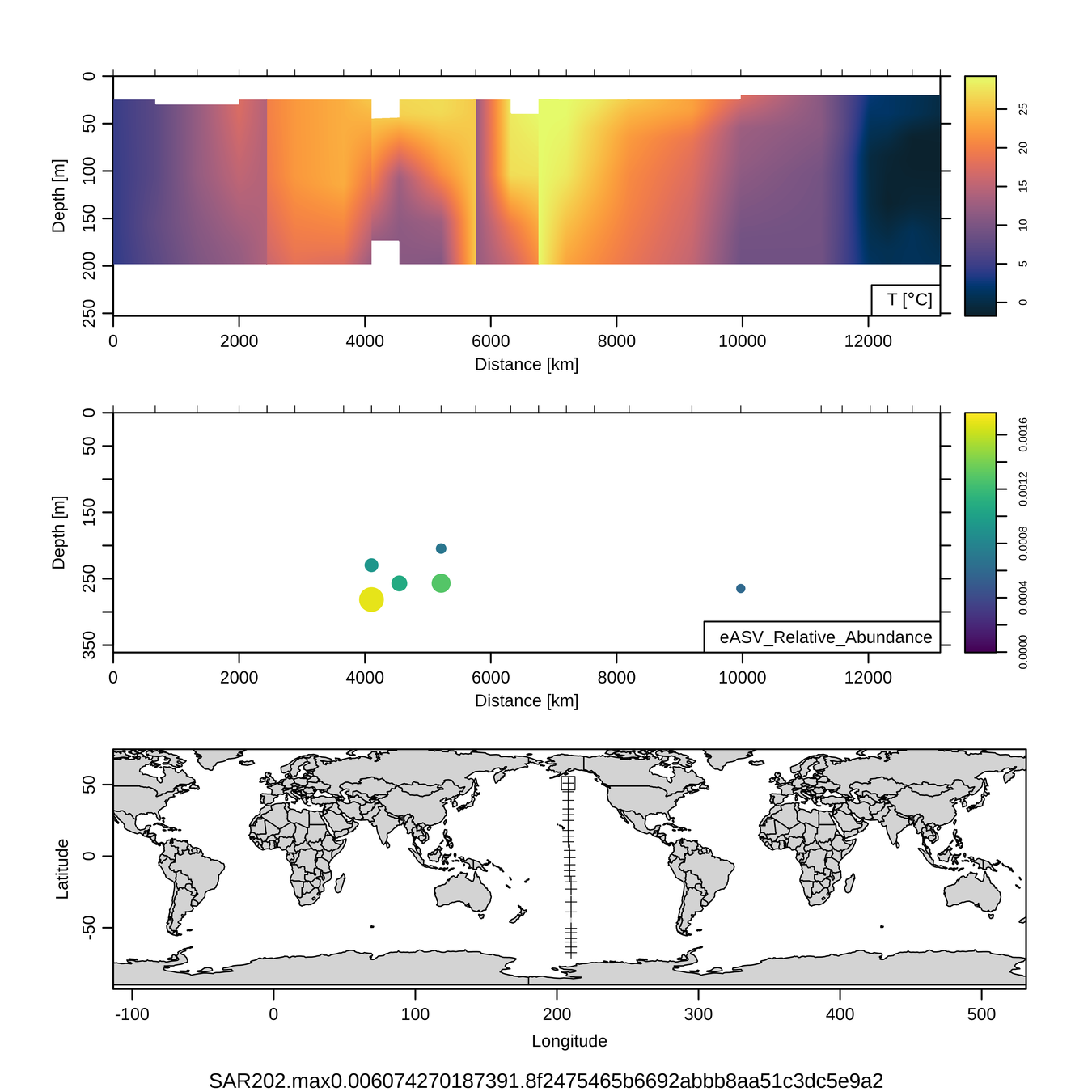
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
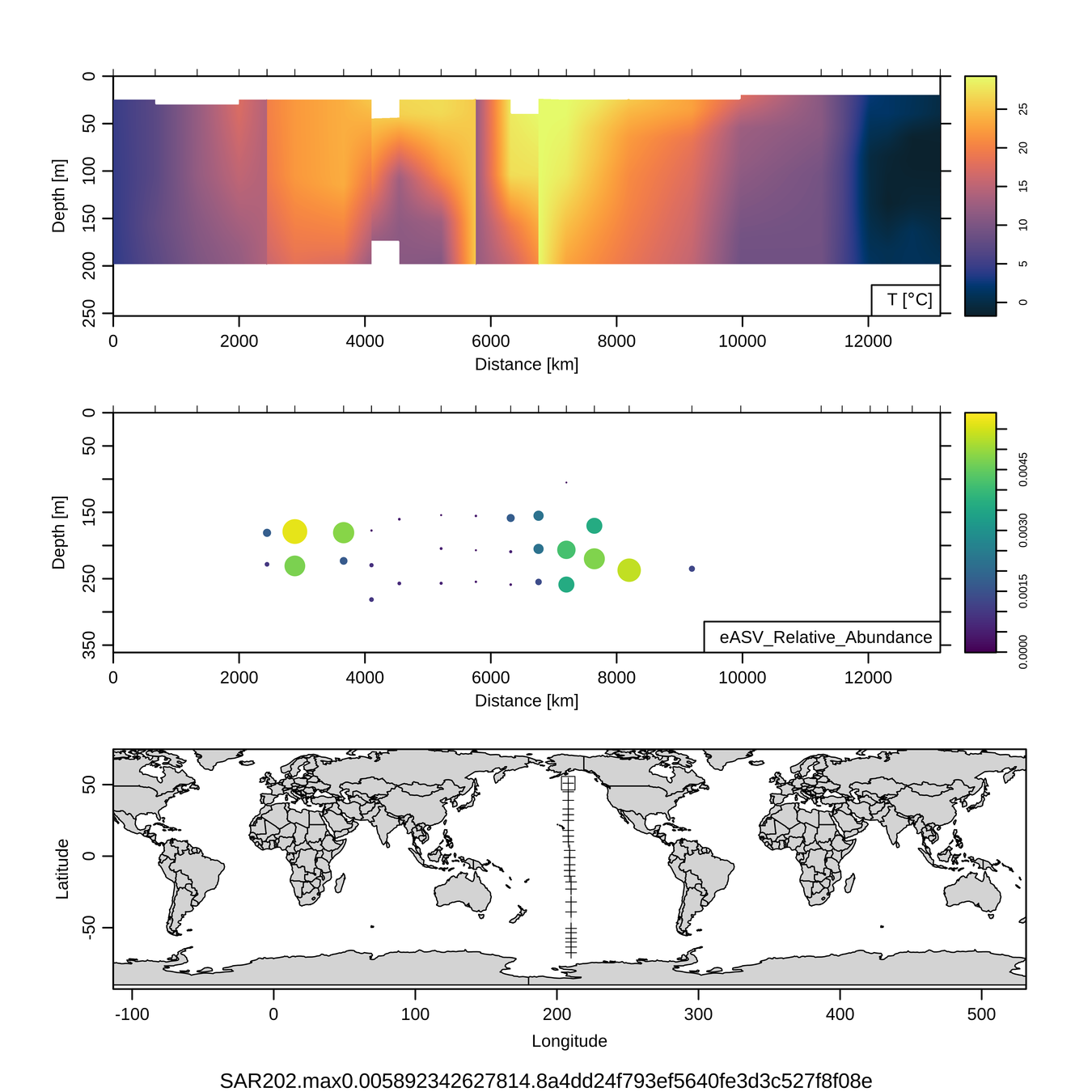
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
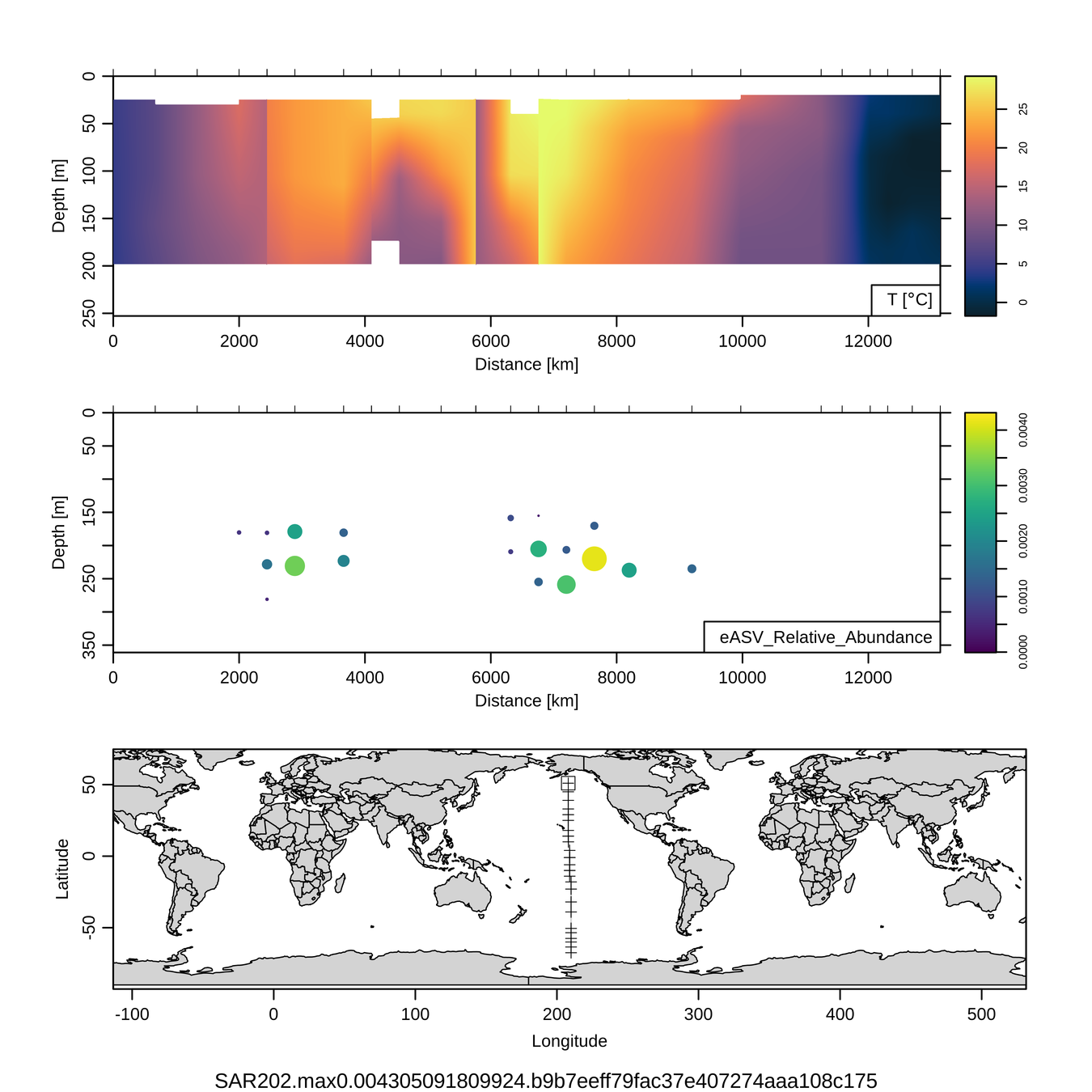
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
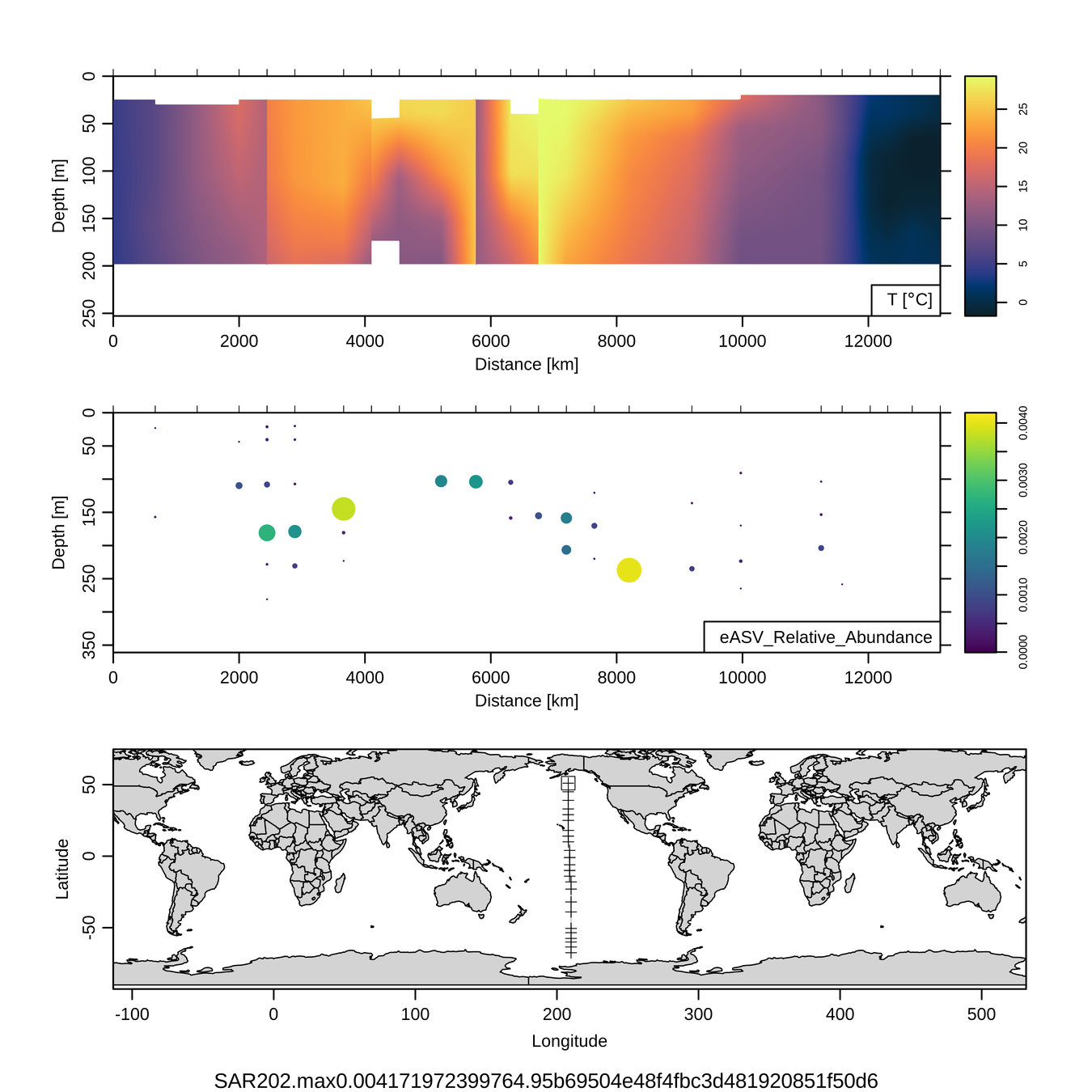
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
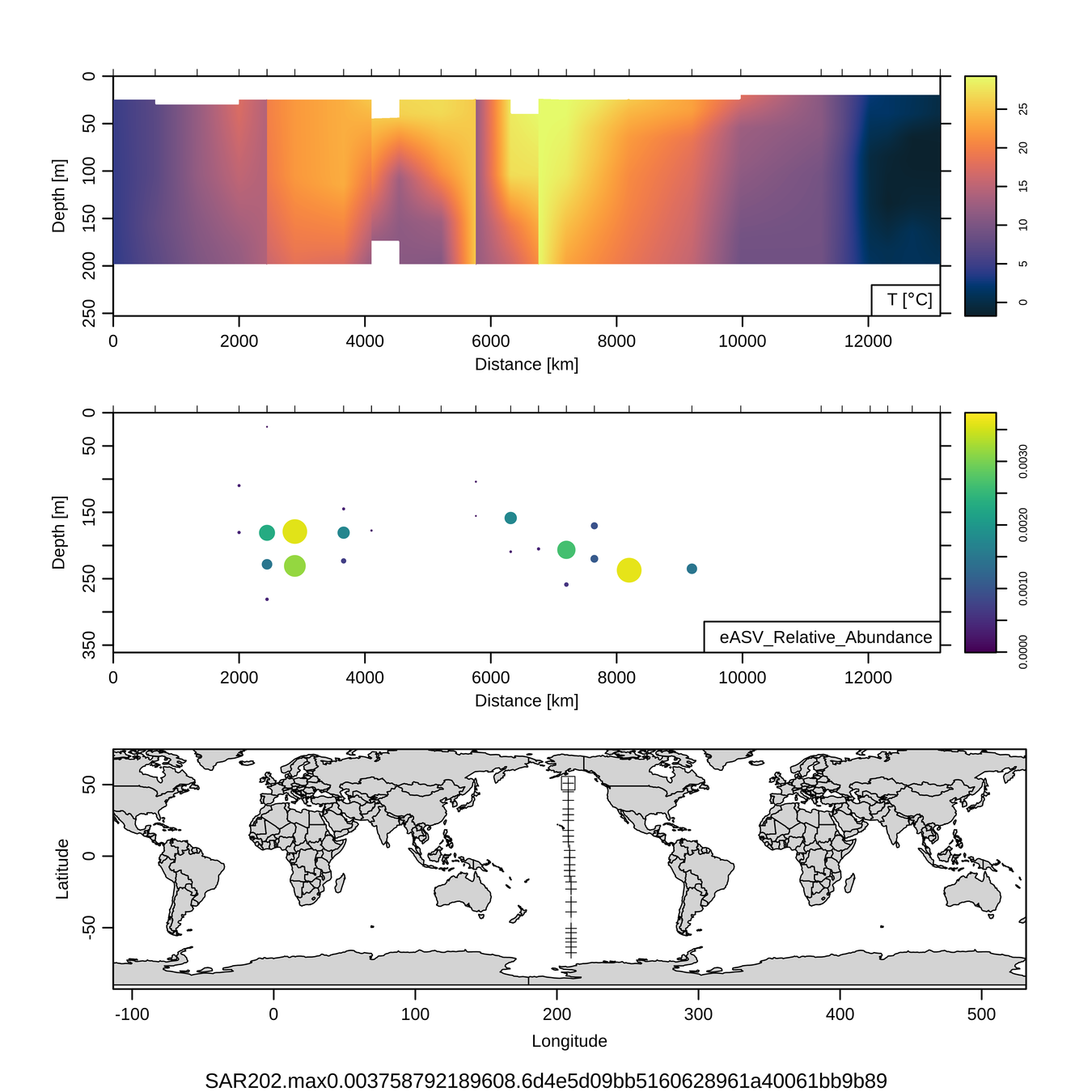
This is only the top 10 from one bacterial taxon!
Similar patterns for phytoplankton (and even some Metazoa)
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"
CBIOMES 2022 Annual Meeting : An Ecosystem Map
By jcmcnch
CBIOMES 2022 Annual Meeting : An Ecosystem Map
- 52



