-
Universal metabarcoding measures the abundance of planktonic life across space / time
- Comprehensive (primers target all cellular life)
- Sensitive with deep sequencing
- Specific with "denoising" algorithms




Microbe art: @claudia_traboni
Taxa-specific activity






"Biobank" (collected from time-series sites / field campagins)
Environmental Sample


Viable cells
DNA
RNA
T/S/Chl./Nut.
Fixed cells
Barcoding

Core environmental data

Projects inspired by environmental data

Cultivation
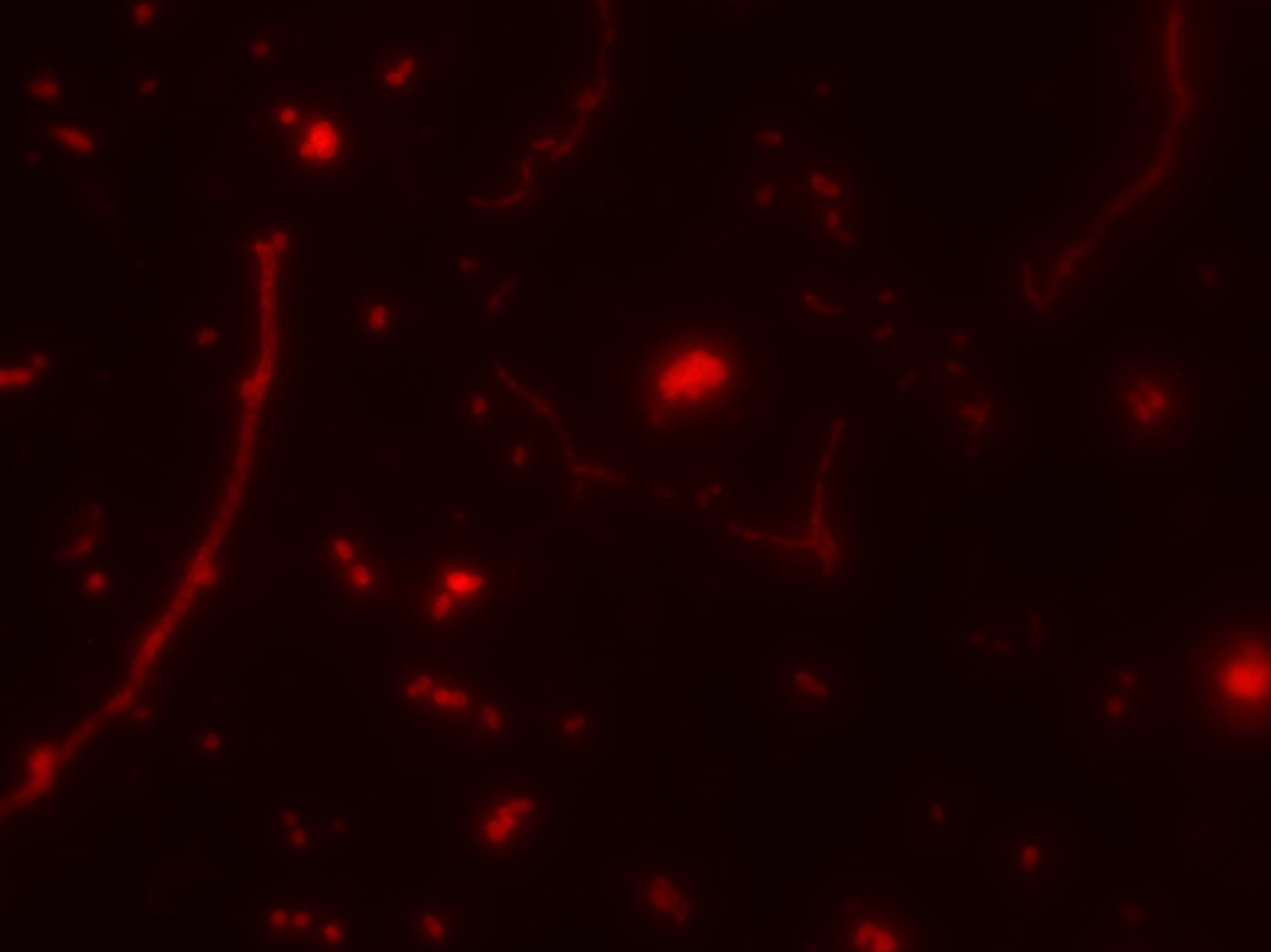
HCR-FISH
'omics


Kalmbach et al. (2017), arXiv:1703.07309v1

- Taxonomy (CARD/HCR-FISH) / transcription (FISH-TAMB)
- Respiration, cell viability (RSG); Polyphosphate content (DAPI)
Automatic cell-sorting + environmental databases = taxon-specific activity or genetic potential measurements
Many uses for sorted cells:
- Taxon-specific isotope uptake
- Mini-metagenomes with few cells
- Cell isolation for cultivation
Pjevac et al (2019) 10.1111/1462-2920.14739
Multiple sorting axes now possible


Microbe art: @claudia_traboni
p16S
e16S
18S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
3-domain metabarcoding quantifies the whole ecosystem

Microbe art: @claudia_traboni
18S
p16S
e16S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
3-domain metabarcoding quantifies the whole ecosystem
Novelty of project:
- Recover information from legacy datasets
- Potentially improve ecological resolution of metabarcodes
- "Rosetta stone" for inter-comparisons, FISH probe design


Dueholm et al. (2020) mBio, e01557-20
Database of full-length 16S rRNA

in silico method optimization
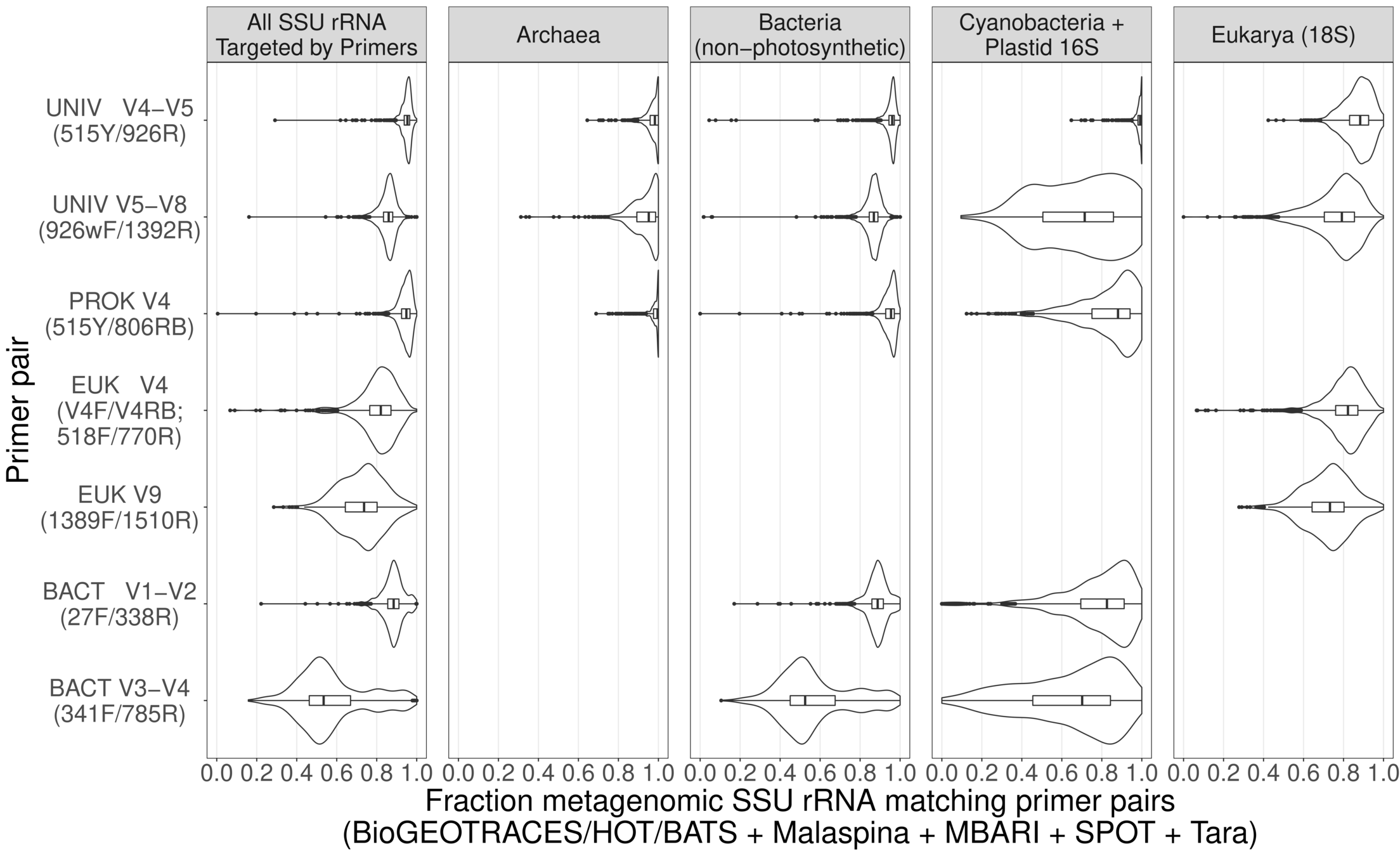
Universal metabarcoding primers work almost perfectly across global oceans
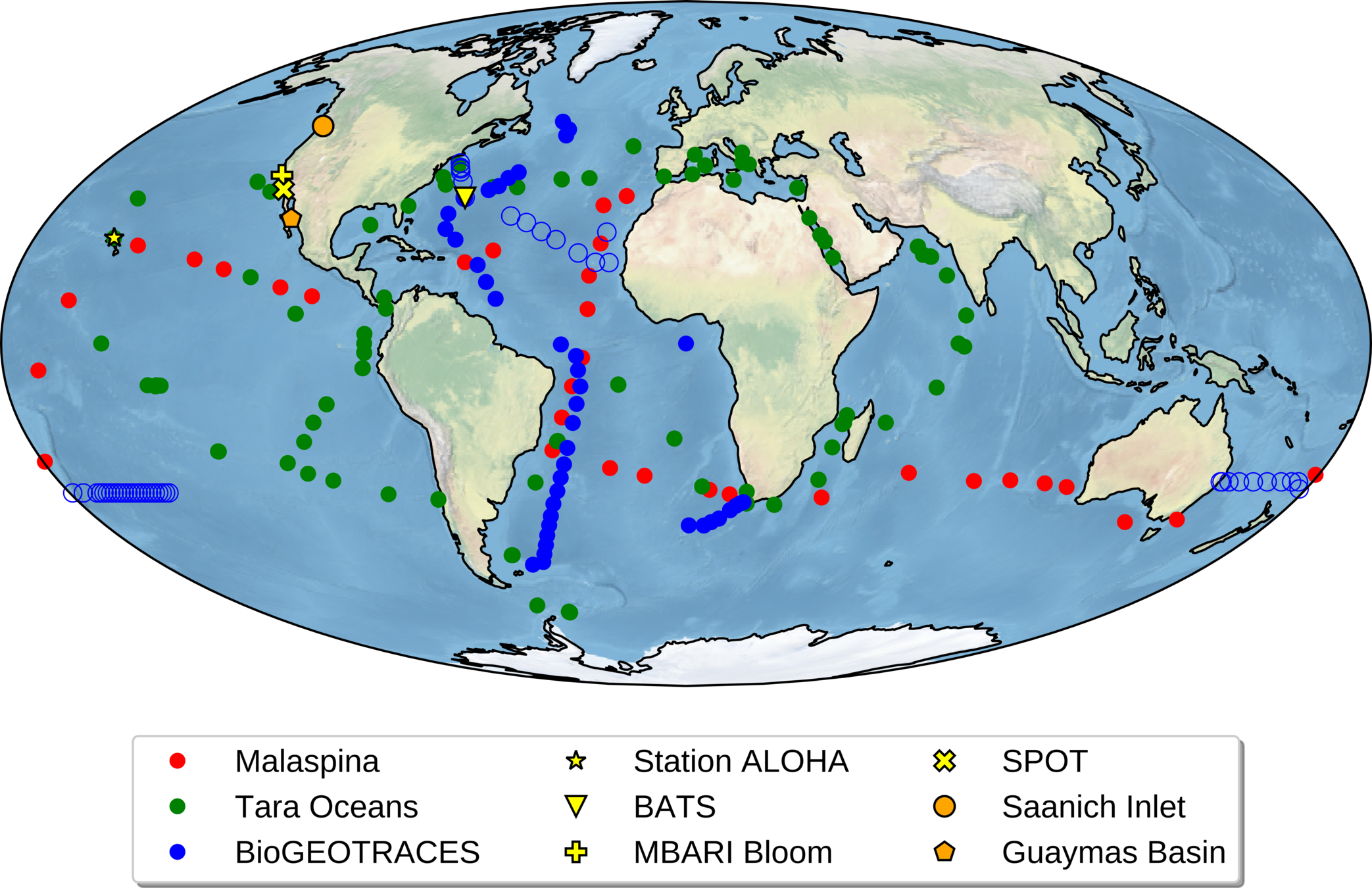
Global metagenomes
plastid
16S
mito 16S
nuclear 18S
Space / time
Abundance
A eukaryotic phytoplankter

"eDNA"
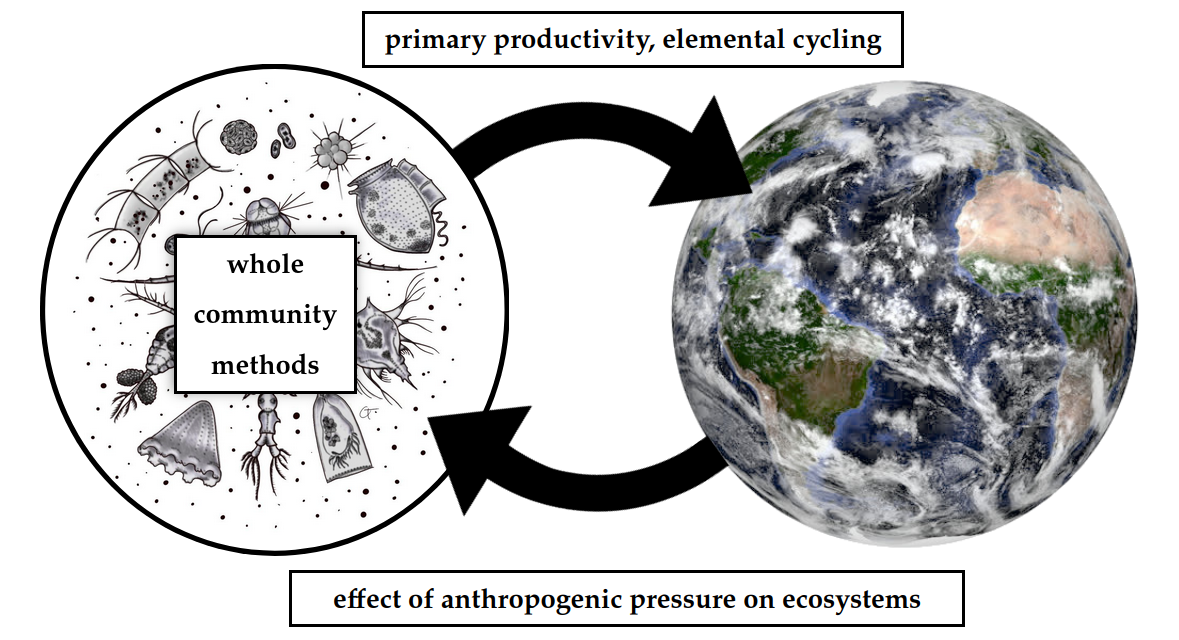
How do microbial communities* influence the Earth System**,
and vice-versa***?

*whole community methods

**primary productivity, elemental cycling

***effect of anthropogenic pressure on ecosystems
Microbe art: @claudia_traboni
My core research question
Microbiology
Bioinformatics
Environmental science
Long term vision: A lab that links microbial diversity & biogeochemical function in the context of global ecosystem change with modern 'omics-enabled techniques
How do microbial communities influence the Earth System, and vice-versa?


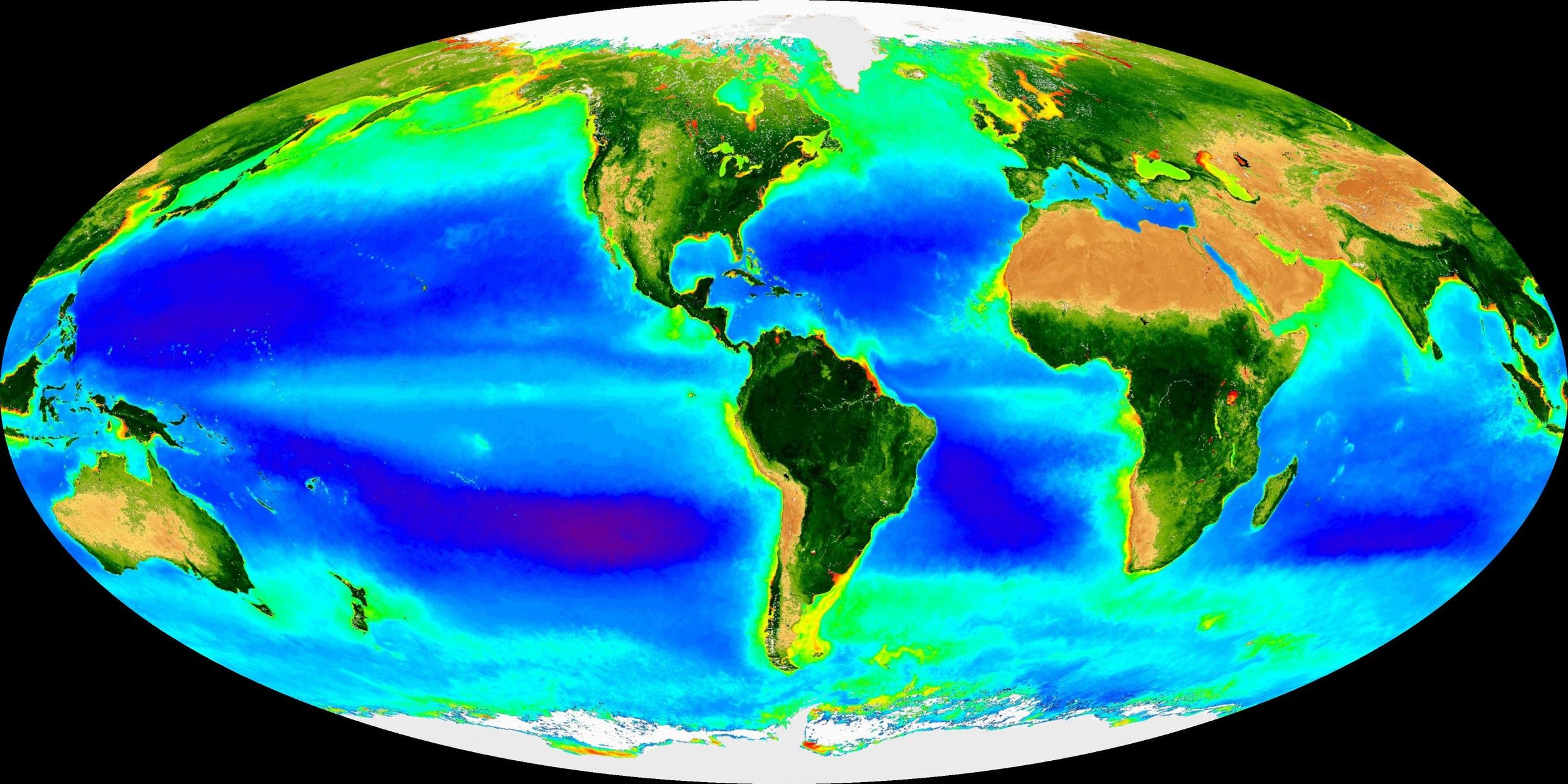


Motivated by desire to understand the Earth System

We anticipate that as trait-based biogeography continues to evolve, micro- and macroorganisms will be studied in concert, establishing a science that is informed by and relevant to all domains of life.” -Green, Bohannan, and Whitaker, Science (2008)
Ecosystem-centered teaching & research
Welcome to the lab!
The StFX Microbial Ecology Lab studies the interactions between microbes (Bacteria, Archaea, and Eukarya), their physical environment, and the biosphere as a whole. We use a combination of field work, modern DNA sequencing, and classical microbiological approaches to address diverse research questions. Check out stfxmicroeco.ca to learn more!
Lab Contact:
Dr. Jesse McNichol
JBB 419
(902) 867-2386
jmcnicho@stfx.ca


Sebastián & Gasol (2019), 10.1098/rstb.2019.0083 ; McNichol et al (2018) 10.1073/pnas.1804351115 ; Zehr (2015) 10.1126/science.aac9752; Bramucci et al (2021) 10.1038/s43705-021-00079-z

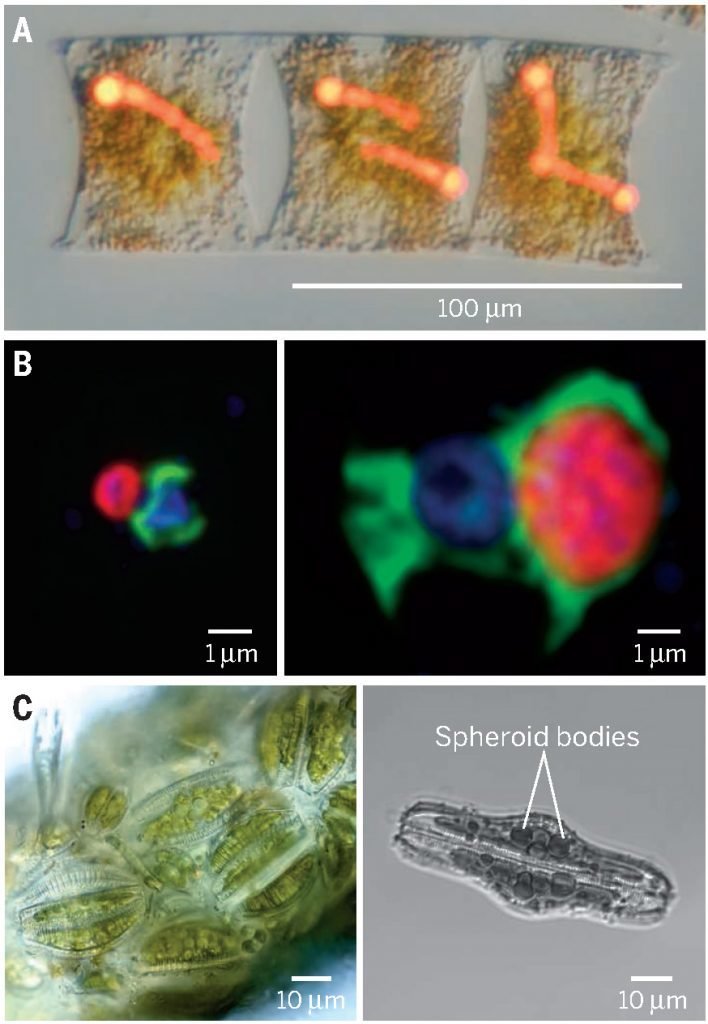

Microscale ecology & biogeochemistry

Slides 4 CBIOMES 2023
By jcmcnch
Slides 4 CBIOMES 2023
- 56



