CBIOMES Annual Meeting (2021)
Jesse McNichol, Postdoc, Fuhrman Lab
University of Southern California


Progress & prospects for using universal primers in modelling work
Single assay for Bacteria, Archaea & Eukarya

Microbe art: @claudia_traboni

"ASVs" = (Exact) Amplicon Sequence Variants


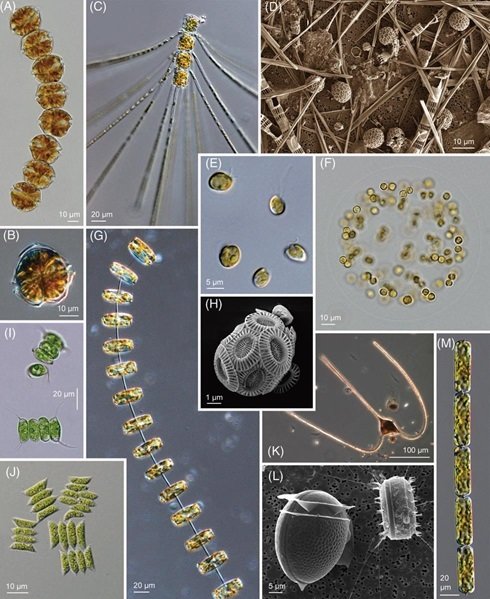
A comprehensive picture of phytoplankton


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


What do relative abundances mean?



Scaled by total chlorophyll (CTD)
An integrative framework

+ Chlorophyll

+ Flow cytometry


Scaled ASV data
r_i^{j} = t_i a^{j} Q_i n_{i}^{j}
Mapping model traits to ASV data
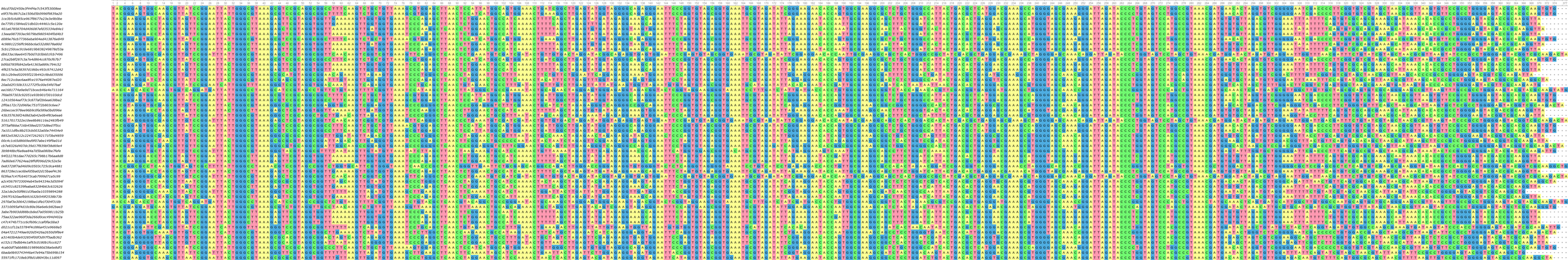
ASV DNA data

ASV phylogeny
Using the full dataset

Worden et al., 2015 (Science)

Full community profile


Single cell genomes from Pachiadaki et al., 2019

Network inference
Future collaboration



pmel.noaa.gov/eoi/staff/resing/clivar.html

Reygondeau et al., 2013
Conclusions


1. New datasets

2. DARWIN - data comparisons

3. Data for collaboration

4. Expanding reach of models
Acknowledgements


Paul Berube & Steven Biller

Yi-Chun Yeh (PhD candidate, Fuhrman lab)

Questions?
Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


Model-data intercomparison (GA03)


CBIOMES2021_eASVs
By jcmcnch
CBIOMES2021_eASVs
- 69