Using Global Metagenomes to Evaluate and Improve PCR Primer Coverage and The Application of 3-Domain Amplicon Data to Trait-Based Models
Jesse McNichol1, Paul M. Berube2, Steven J. Biller2,3,
Sallie W. Chisholm2, Jed A. Fuhrman1
1University of Southern California
2Massachusetts Institute of Technology
3Wellesley College


Image: @claudia_traboni (PhD candidate, Insituto Ciencias Marinas CSIC)
How do we understand this whole system?
Models do well for phytoplankton
http://darwinproject.mit.edu
How well do models represent reality? How to add heterotroph taxa?

-
Universal Ribosomal PCR Primers target the SSU rRNA of (almost) everything:
- Quantify the whole cellular community from A-Z (Archaea + Bacteria + Organelles + Protists + Zooplankton)
- 16S and 18S on same scale - ratios meaningful
- Low cost (~$8/sample, 200,000 sequences/ea)
- Amplicon sequence variants (ASVs) allow mapping, intercomparison
Universal primers as a mapping tool
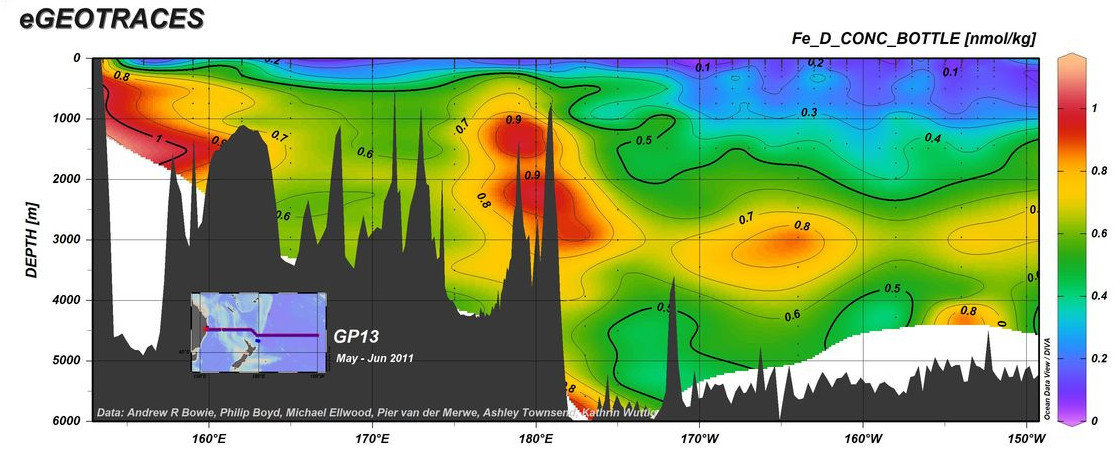
https://www.egeotraces.org/sections/GP13_Fe_D_CONC.html

@claudia_traboni
-
Detailed protocols available on https://fuhrman-lab.github.io
- Lab protocol
-
in-silico workflow:
- Splits 16S and 18S automatically (analyzed separately)
- Denoises with DADA2/deblur in qiime2 => ASVs
- Splits data automatically into categories (e.g. chloroplasts)
- Step-by-step guide on protocols.io, qiime2 visualizations
Universal primers are easy to analyze!



Universal primers complement existing techniques
Universal primers complement existing techniques
- Metagenomic validation of universal primer quantification and environmental coverage
- Use cases for universal primer data (with BioGEOTRACES data)
- Future work
Talk Outline

https://www.egeotraces.org/sections/GP13_Fe_D_CONC.html
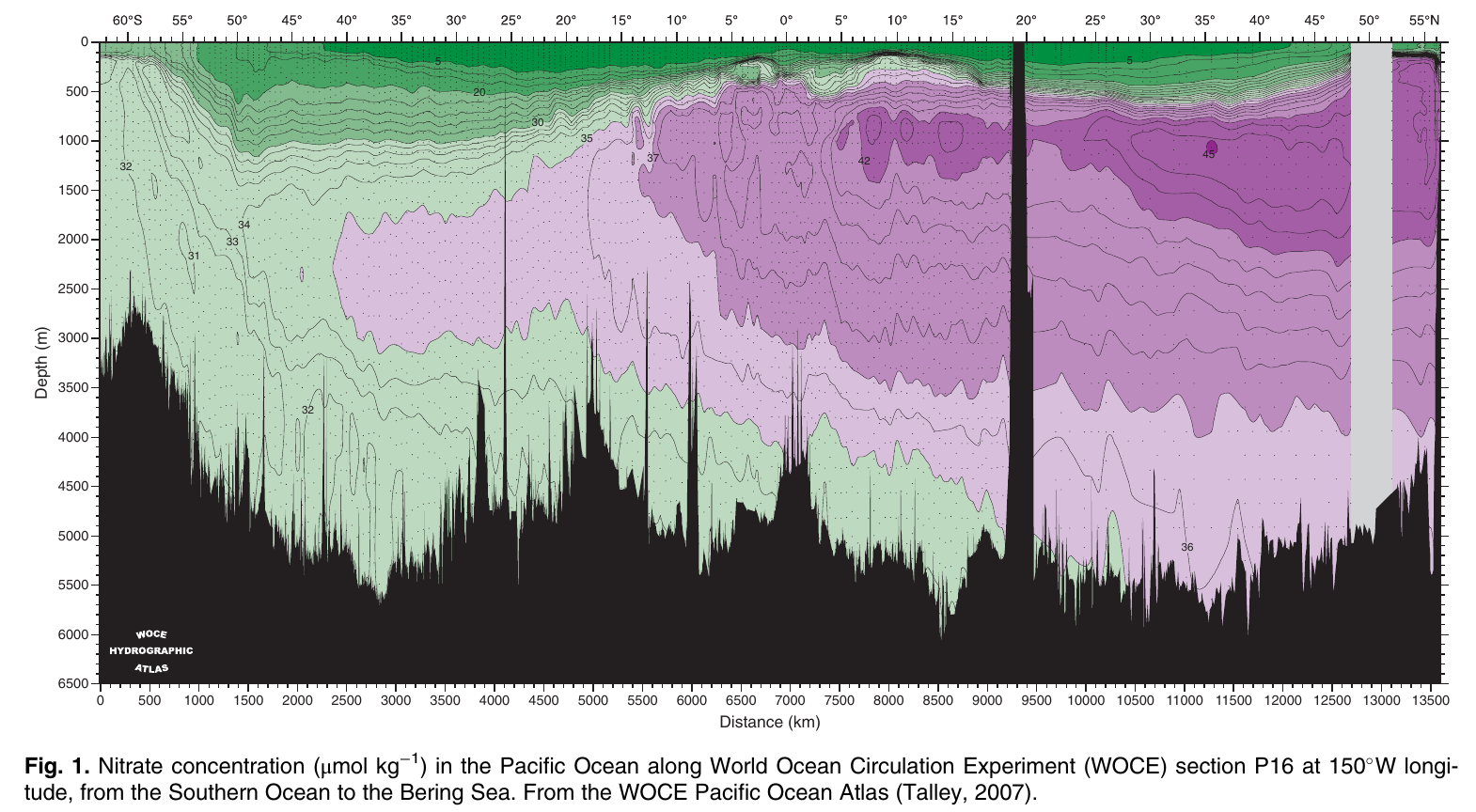
Universal primers are quantitative




Metagenome
abundances
(16S)
PCR amplicon
abundances (16S)
Also quantitative "in the wild"
Similarly quantitative "in the wild"; 18S less so
Metagenome
abundances
(18S)
PCR amplicon
abundances (18S)

Similarly quantitative "in the wild"; 18S less so
Metagenome
abundances
(18S)

PCR amplicon
abundances (18S)
What about primer bias?

Inspired by this paper, we are using metagenomes to determine theoretical primer performance across ecosystems
MGPrimerEval : a reproducible snakemake pipeline
- Compares primers to unassembled MG reads
- tinyurl.com/titus-workflow-philosophy
- Reproducible by conda/docker integration
- github: tinyurl.com/MGPrimerEval
https://www.physalia-courses.org

Sorting, aligning, subsetting

Primer-MG comparisons, data summary

QC, SSU rRNA sifting, repeat removal
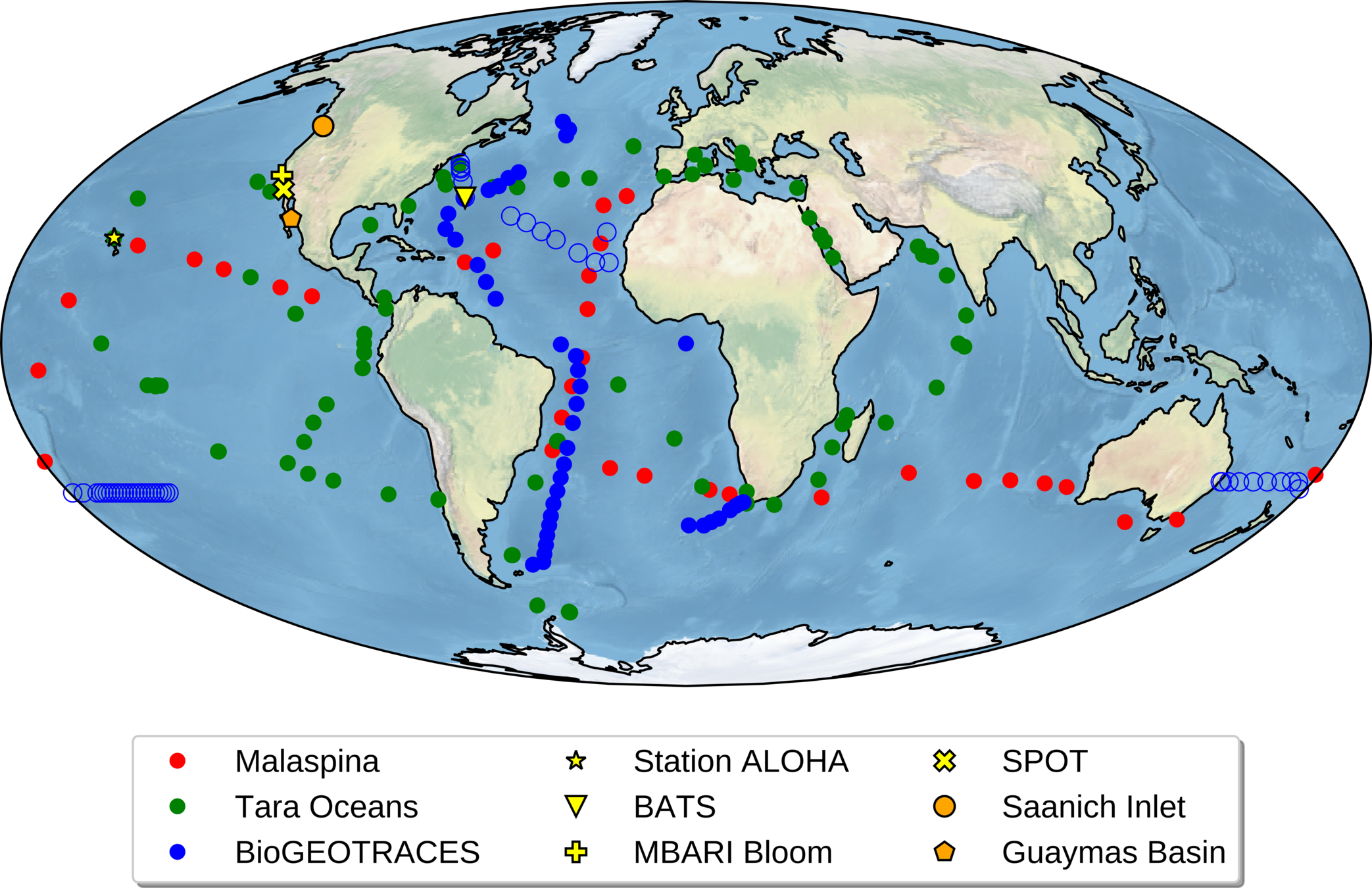
MGPrimerEval : datasets
-
TARA oceans, BioGEOTRACES, MBARI bloom MG, Malaspina, SPOT/Catalina MG
- Total = 1,218 metagenomes
- Globally distributed, mostly oxic, pelagic samples
Results: fraction of MG perfectly matching primer
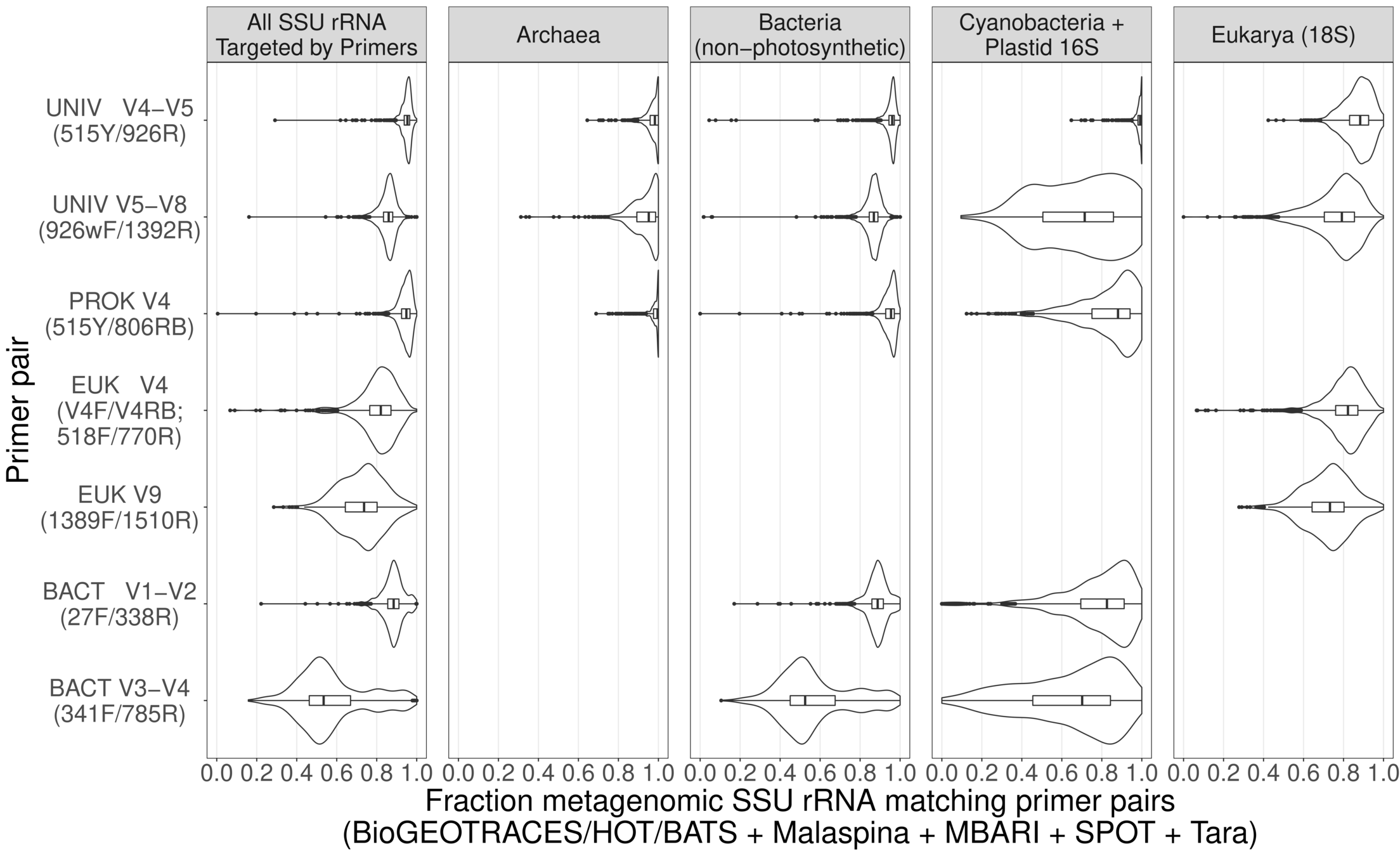

Conclusions
- Our primers match vast majority of MG sequences perfectly
- Nearly 100% for 16S, ~90% for 18S
- The glass is also half full! Existing primers can be improved
- Optimize coverage for biogeochemically-important taxa


preprint:
tinyurl.com/oceanprimers
- Metagenomic validation of universal primer quantification and environmental coverage
- Use cases for universal primer data (with BioGEOTRACES data)
- Future work
Talk Outline

https://www.egeotraces.org/sections/GP13_Fe_D_CONC.html
2. Use cases for Universal Primers:
a) Resolution
b) Sensitivity (detection limit)
c) Modelling
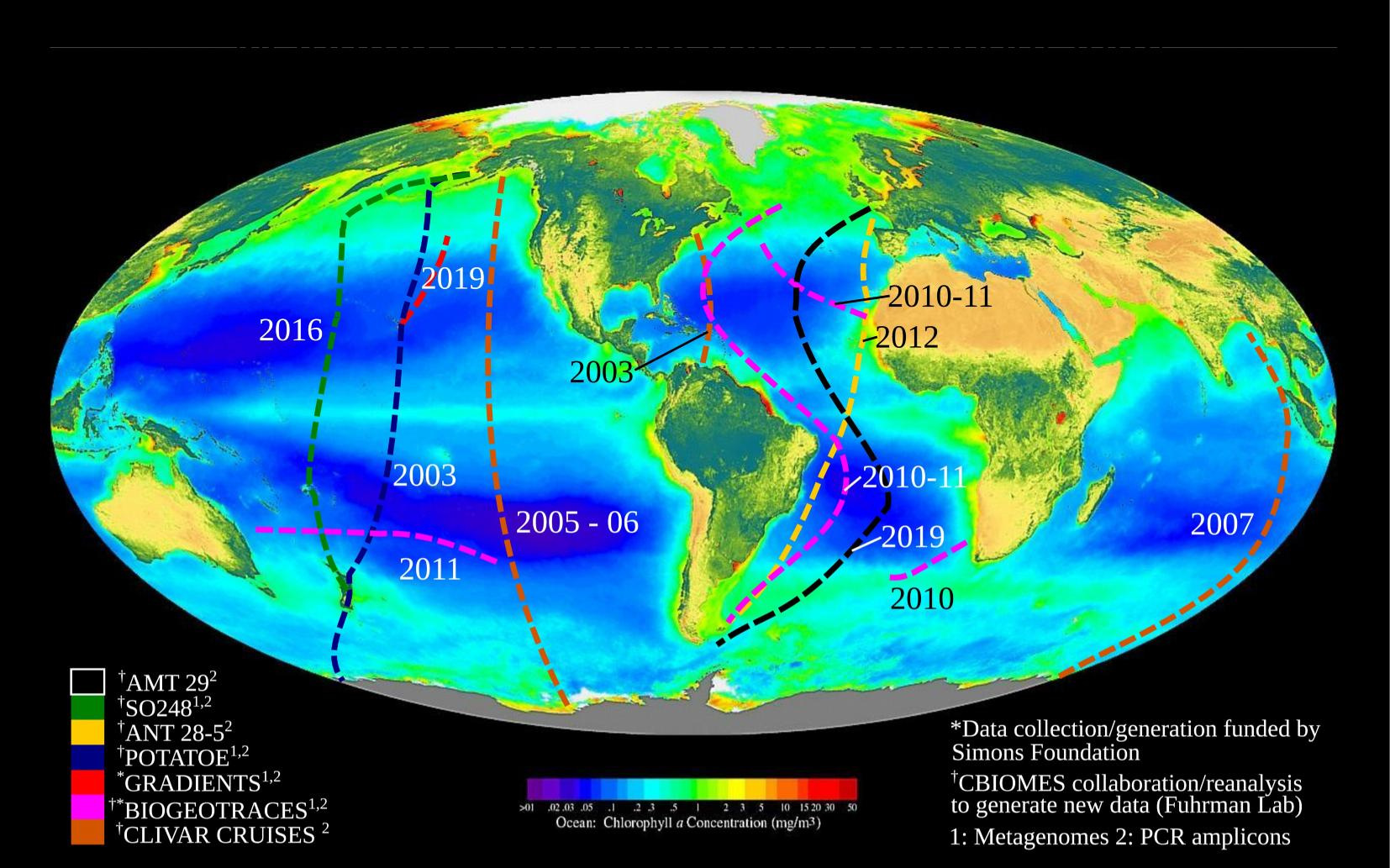
Data source: BioGEOTRACES amplicons


Will present new data from these transects
These are "whole water" samples (> 0.2 µm)
Universal primers + whole water samples =
comprehensive profile
a) Resolution
- Are short amplicons (~373 bp) enough to resolve ecologically-relevant subgroups of Prochlorococcus?
b) Sensitivity (detection limit)
c) Modelling
2. Use cases for Universal Primers:
Majority of ASVs consistent with genome phylogeny and associated with a particular ecotype
*Berube et al. eLife 2019;8:e41043.
Resolution: Prochlorococcus ASVs



ASV hits to clade
Resolution: Prochlorococcus ASVs

HLII clade ASV
Higher abundance in warmer waters

HLI clade ASV
Abundance increases as temperature drops
Both distributions consistent with physiological data from pure cultures and field data
Resolution: Prochlorococcus ASVs

Biller et al., 2015 Nat. Rev. Microbio.
Resolution
- Are short amplicons (~373 bp) enough to resolve ecologically-relevant subgroups of Prochlorococcus?
Sensitivity (detection limit)
- Can we sequence deeply enough to find rare, but biologically-relevant organisms like diazotrophs?
Modelling
2. Use cases for Universal Primers:
Sensitivity: Diazotrophs
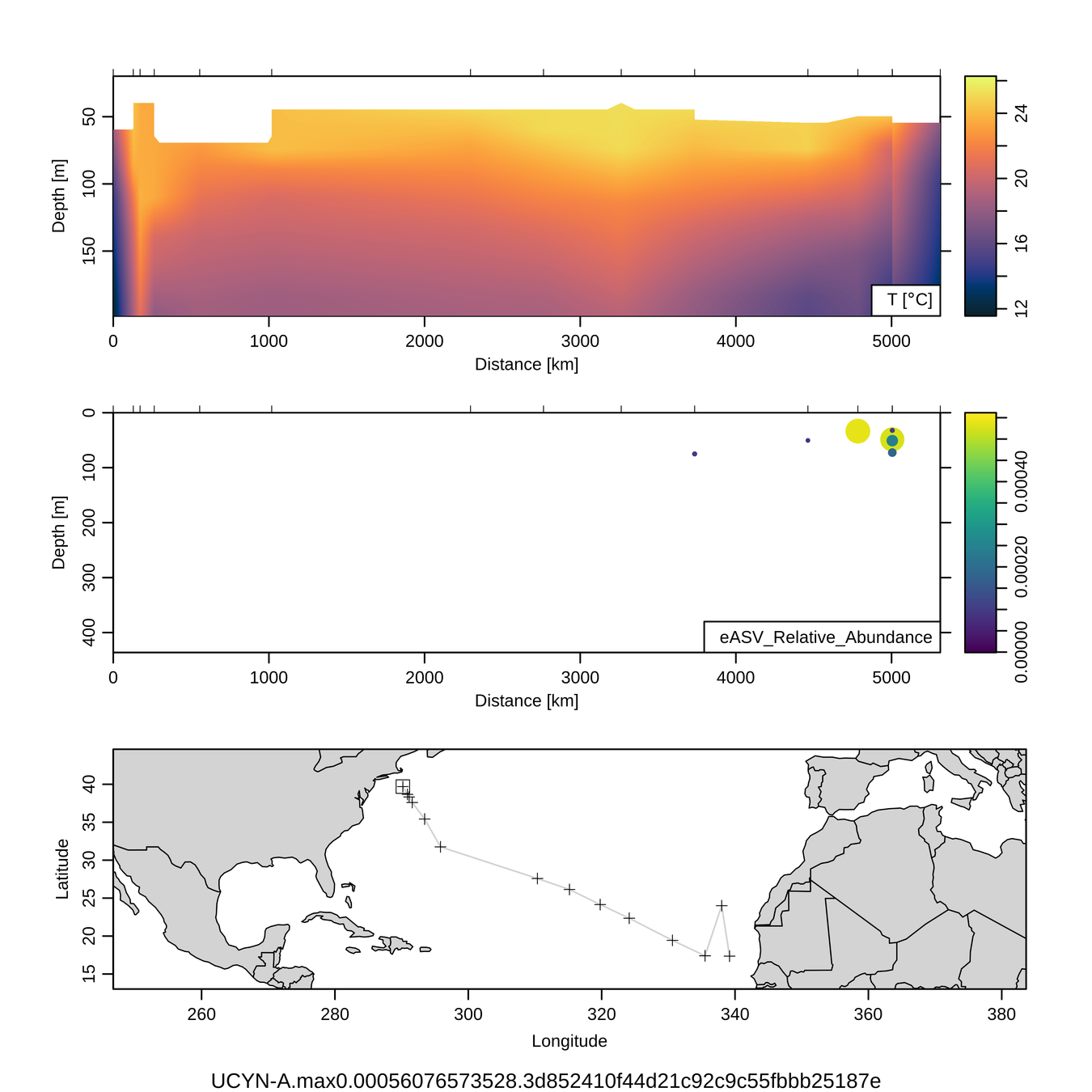
UCYN-A (diazotroph)
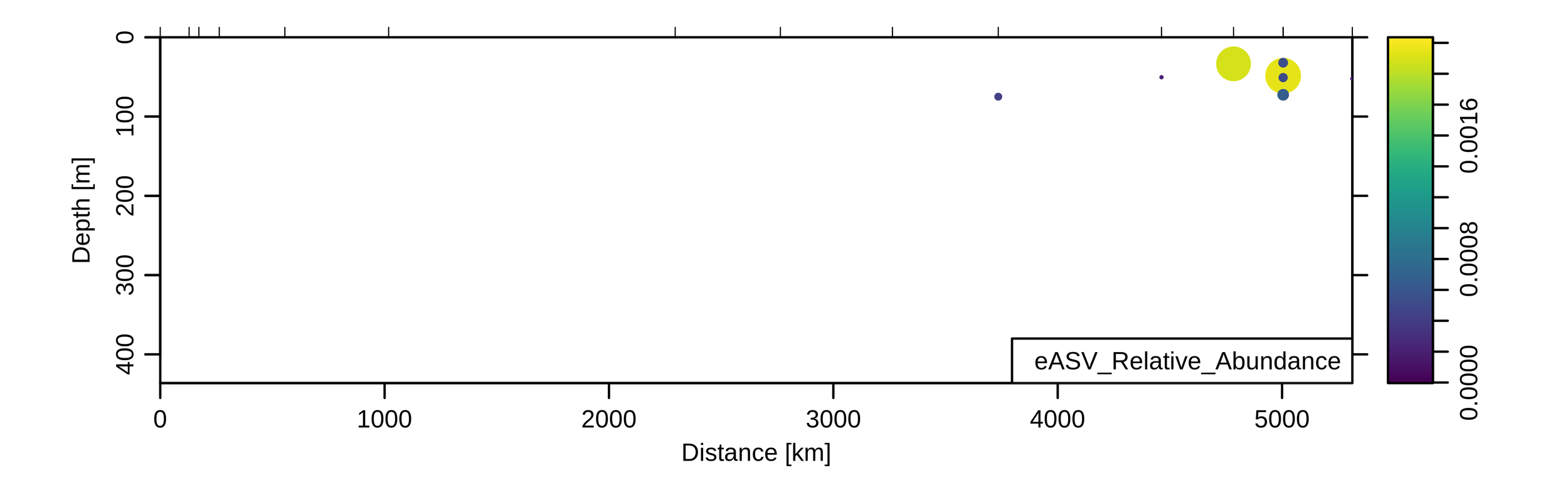
Braarudosphaera chloroplast (putative host)
1/2000
to
1/13,000
Zeroes meaningful
No UCYN-A observed out of
~50,000 reads

UCYN-A (diazotroph)

Braarudosphaera chloroplast (putative host)

qPCR (nifH) data
Sensitivity: Diazotrophs
UCYN-A - Braarudosphaera correlation

UCYN-A (diazotroph)

Braarudosphaera chloroplast (putative host)

Resolution
- Are short amplicons (~373 bp) enough to resolve ecologically-relevant subgroups of Prochlorococcus?
Sensitivity (detection limit)
- Can we sequence deeply enough to find rare, but biologically-relevant organisms like diazotrophs?
Modelling
- How can data be used to improve modelling efforts?
2. Use cases for Universal Primers:
Modelling: Rationale
- Microorganisms process up to 50% of PP
- Not yet included in DARWIN model
-
Amplicons can constrain microbial biogeography
- Use to inform and test modelling approaches

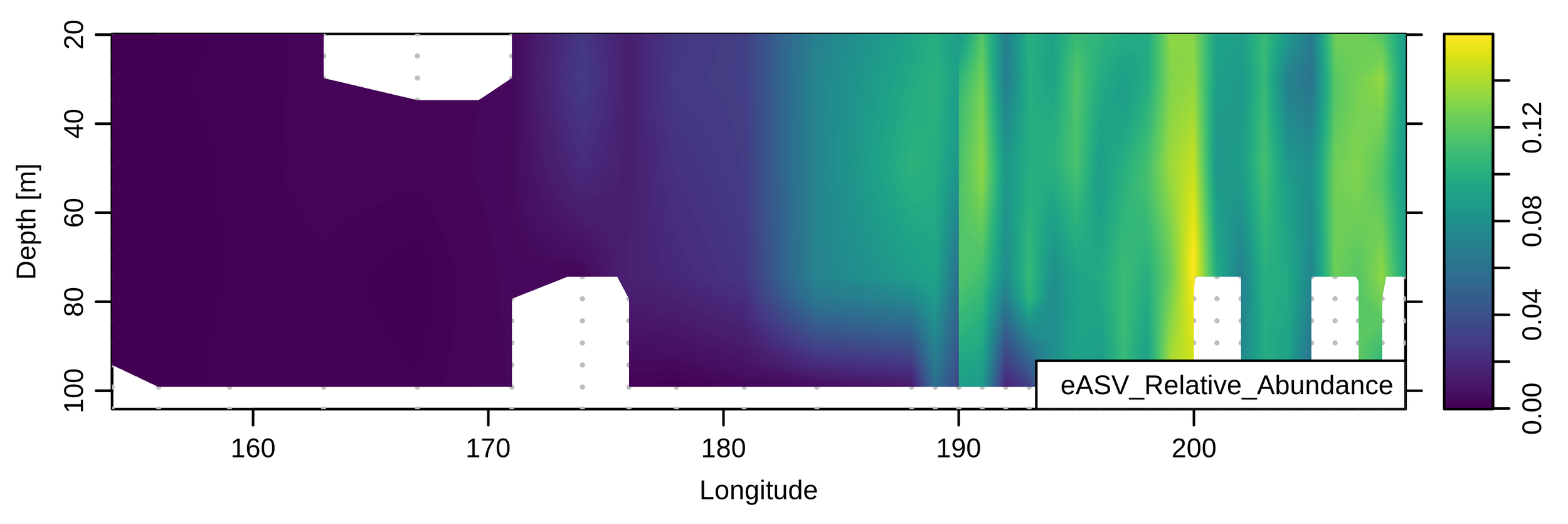
Depth profiles from KN204-stn3
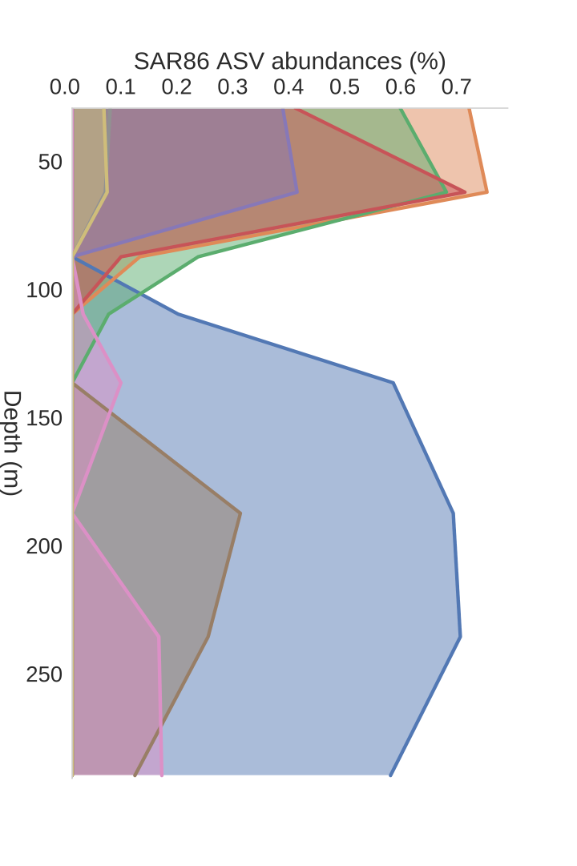
Modelling: ASV (~species) depth profiles
Marine actinobacterium
Extreme oligotroph
Very small size

SAR86, uncultured
Lipid/protein degrader?
Deep/shallow ecotypes?

Marine group II archaea
Also lipid protein/degrader?
Particle attached?

Modelling: Aggregated profiles


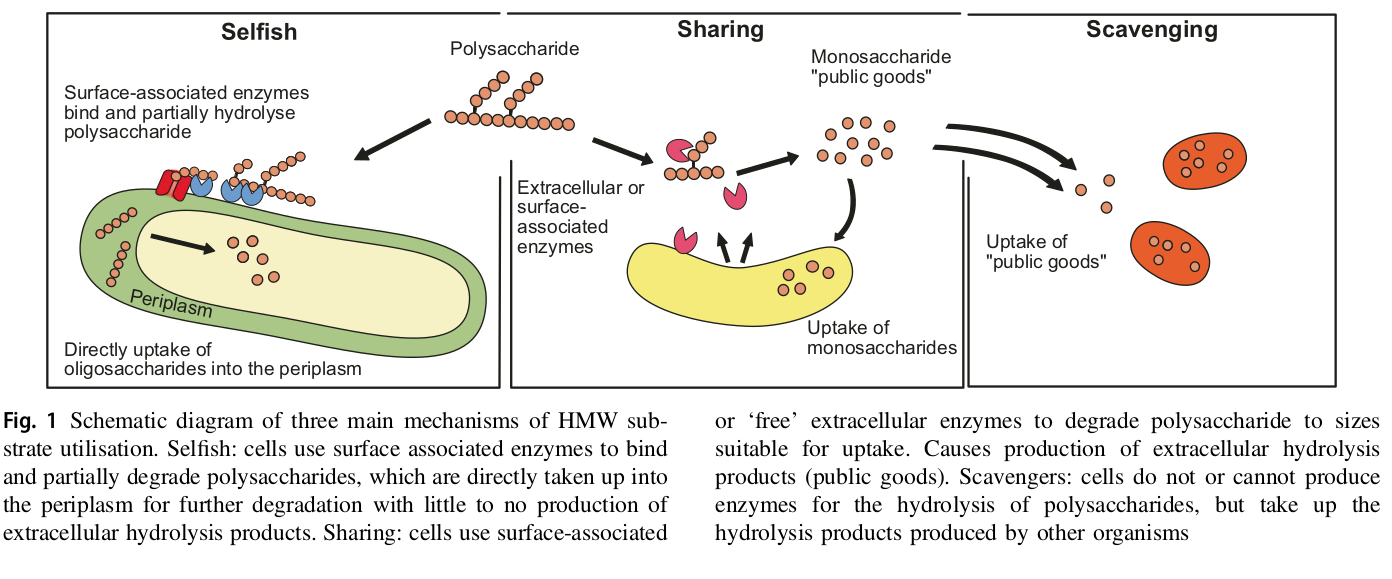
Reintjes et al., 2018, ISME doi.org/10.1038/s41396-018-0326-3



Flavobacteria
Alteromonas
SAR11
Modelling: Defining traits
Biogeography
(e.g. GA03)
Meta'omics (e.g. Iverson et al 2012, Pachiadaki et al., 2019)

Models (e.g. Emily Zakem's work on heterotrophs)
Modelling: Integrating different data types
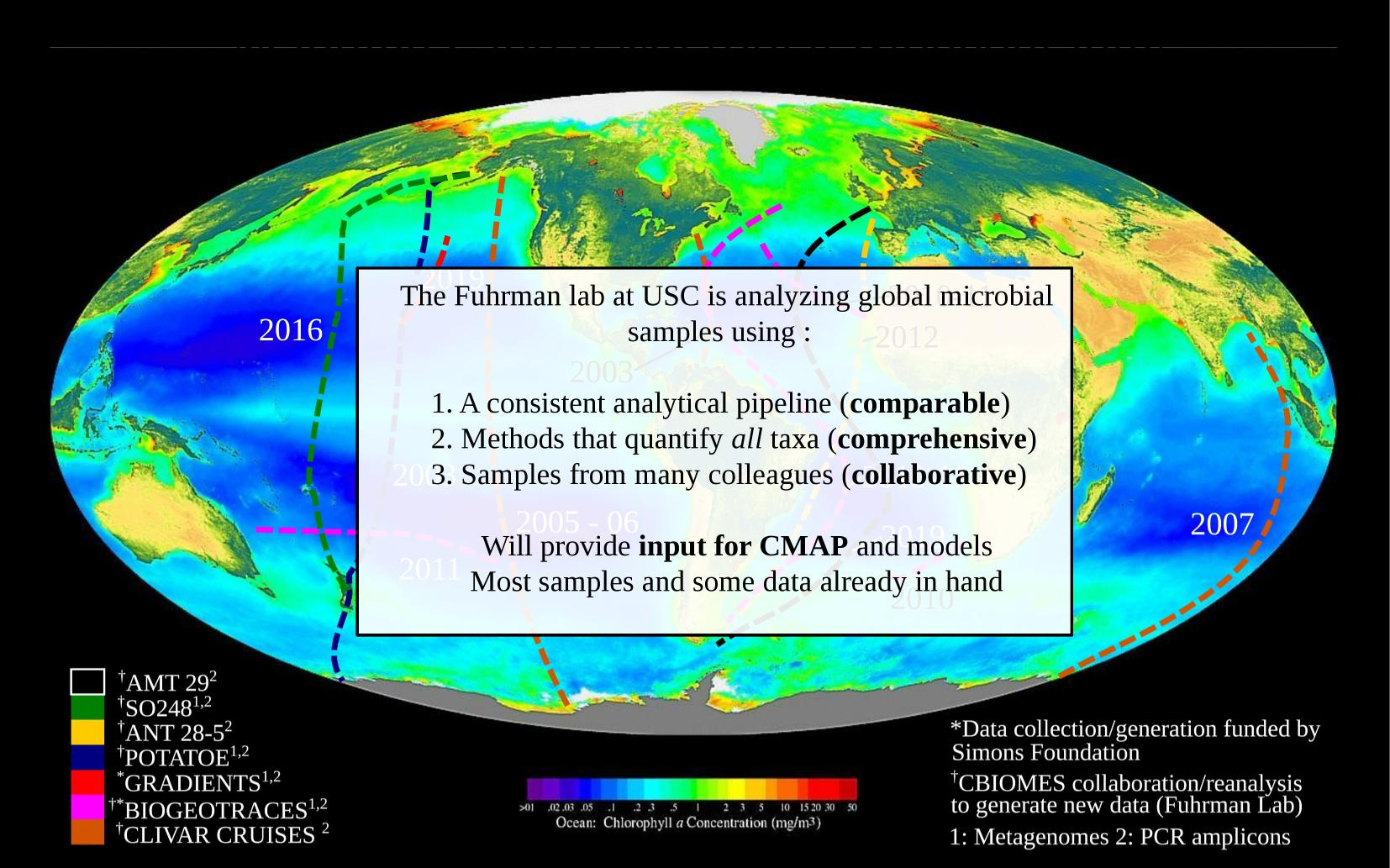
3. Future work: Expanding ocean coverage
- Simons Collaborative Marine Atlas Project (cmap.readthedocs.io)
- Unify data retrieval and co-localization
- ASVs are stable, and make sense in database
Future work: Public access to data in CMAP

Conclusions: Universal Primers
- Cost-effective tool to map whole cellular community from A-Z (Archaea + Bacteria + Organelles + Protists + Zooplankton)
- Guide more focused studies of same DNA (e.g. meta'omics)
- Straightforward to process and interpret
- Can provide guideposts for modelling efforts

@claudia_traboni

https://www.egeotraces.org/sections/GP13_Fe_D_CONC.html
Acknowledgements
- Yi-Chun Yeh (for collaboration on amplicon analysis)
- Shengwei Hou (for help and inspiration on computational analyses)
- Clark Richards (for help with R "oce")
- Ken Youens-Clark (for help with C-microbe-MAP)
- Emily Zakem (for helping me understand modelling)
- Mike Lee (for help separating 16S and 18S in silico)
-
Fuhrman lab crew
- fuhrman-lab.github.io
- tinyurl.com/MGPrimerEval



NB: 18S usually small fraction of total
Will be a smaller % for amplicons
# of 18S sequences recovered will be limited for some regions

Most metagenomes have < 20% 18S
% Eukaryotic amplicons
-
GP13 (mostly oligotrophic surface waters)
- AVG = 3.2% (range: ~1-28%)
- For 200,000 sequences = 6400 18S sequences (2000 - 56,000)
-
GA03 (mostly oligotrophic, more deep-sea sampling)
- AVG = 2.7% (range: 0.17-17%)
DADA2 vs deblur (ASVs vs. MG)

DADA2 vs deblur (ASV vs. ASV)

3. Future work: Expanding ocean coverage
ICBM e-seminar
By jcmcnch
ICBM e-seminar
Zoom seminar to ICBM community Nov 25th 2020
- 50



