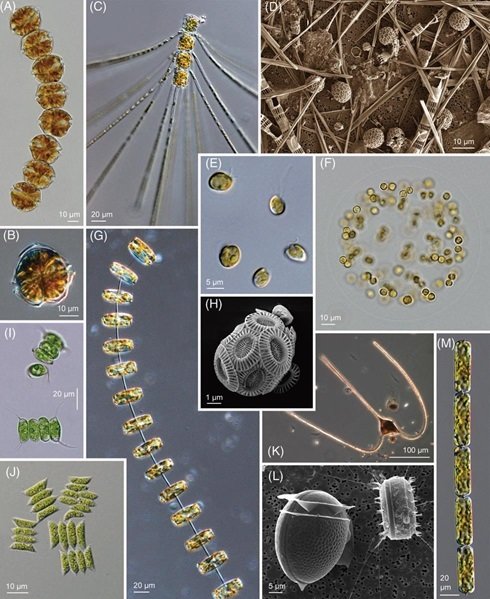
New Datasets for Marine Macroecology:
Metabarcoding, Metagenomics, & Related Techniques
2023-06-21, CBIOMES Annual Meeting
Jesse McNichol

- Why do we have new datasets?
- Method definitions, pros and cons
- Global metagenomes
- Global metabarcodes
- Data & directions
Outline
(1) Cheap DNA sequencing = new datasets
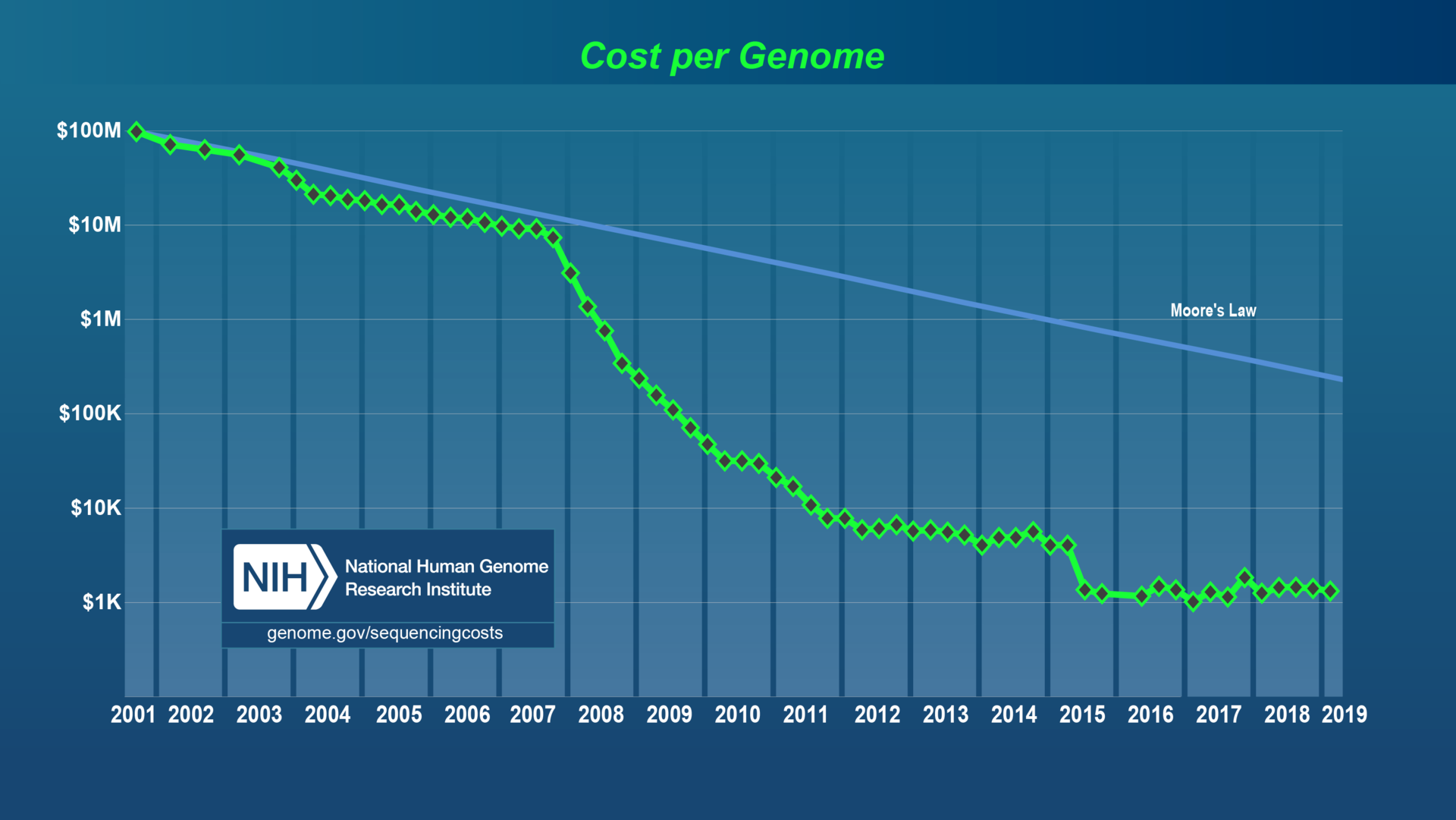
Early sequencing
"Next Generation Sequencing" (NGS)
"3rd generation" sequencing
(1) Different stages

Exploration, discovery
Intercalibration, harmonization
Methodological validation

Ecosystem
Ecosystem

Metagenomics

Metabarcoding
Metabarcoding
(2) Metabarcodes vs metagenomes

Different primers, different regions of barcode
Different primers, different organismal range
Universal
Universal Prok
Universal Euk
Universal Bacteria
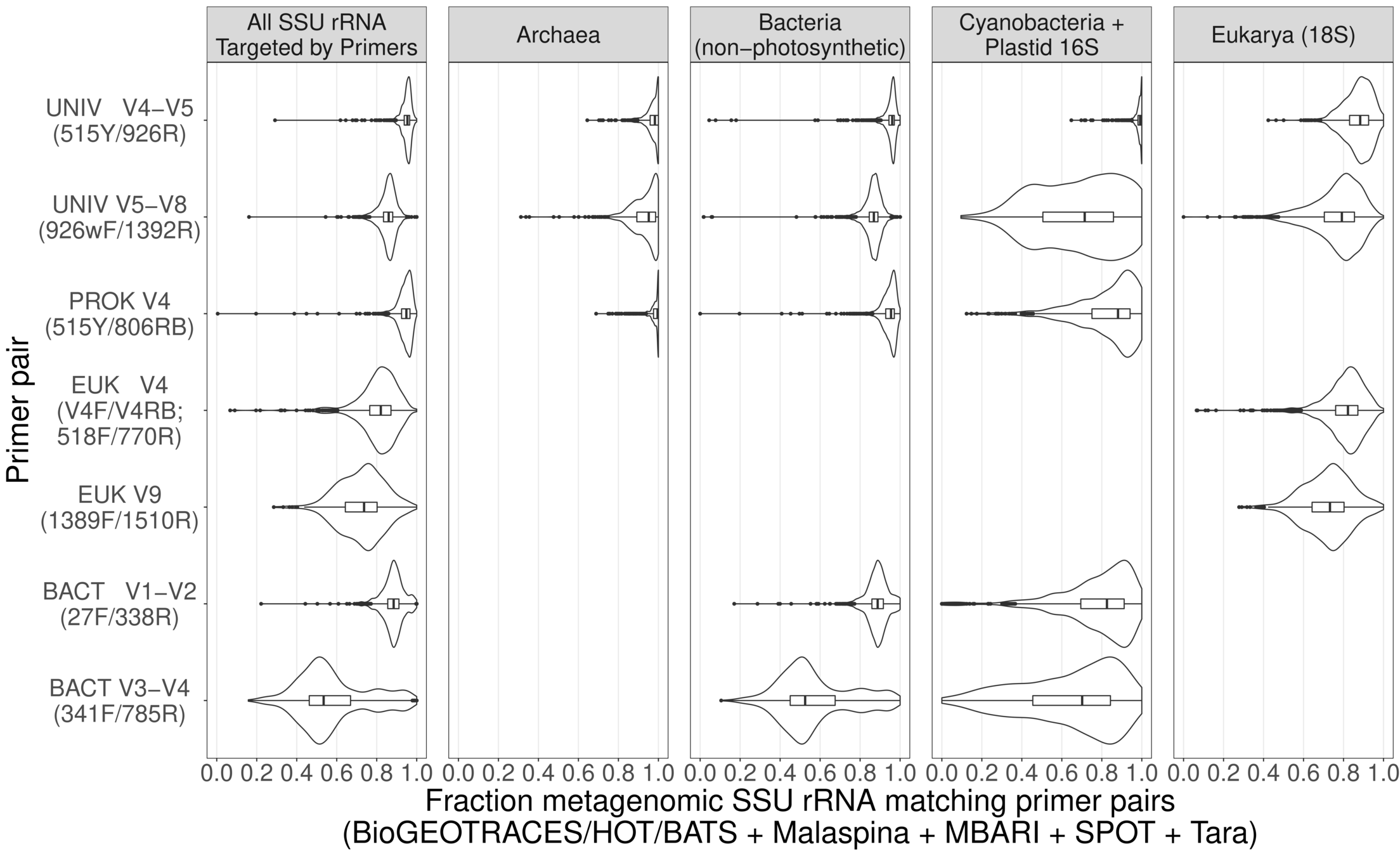

(2) Challenges of metabarcoding
(3) Global shotgun metagenomics = the answer?

BioGEOTRACES
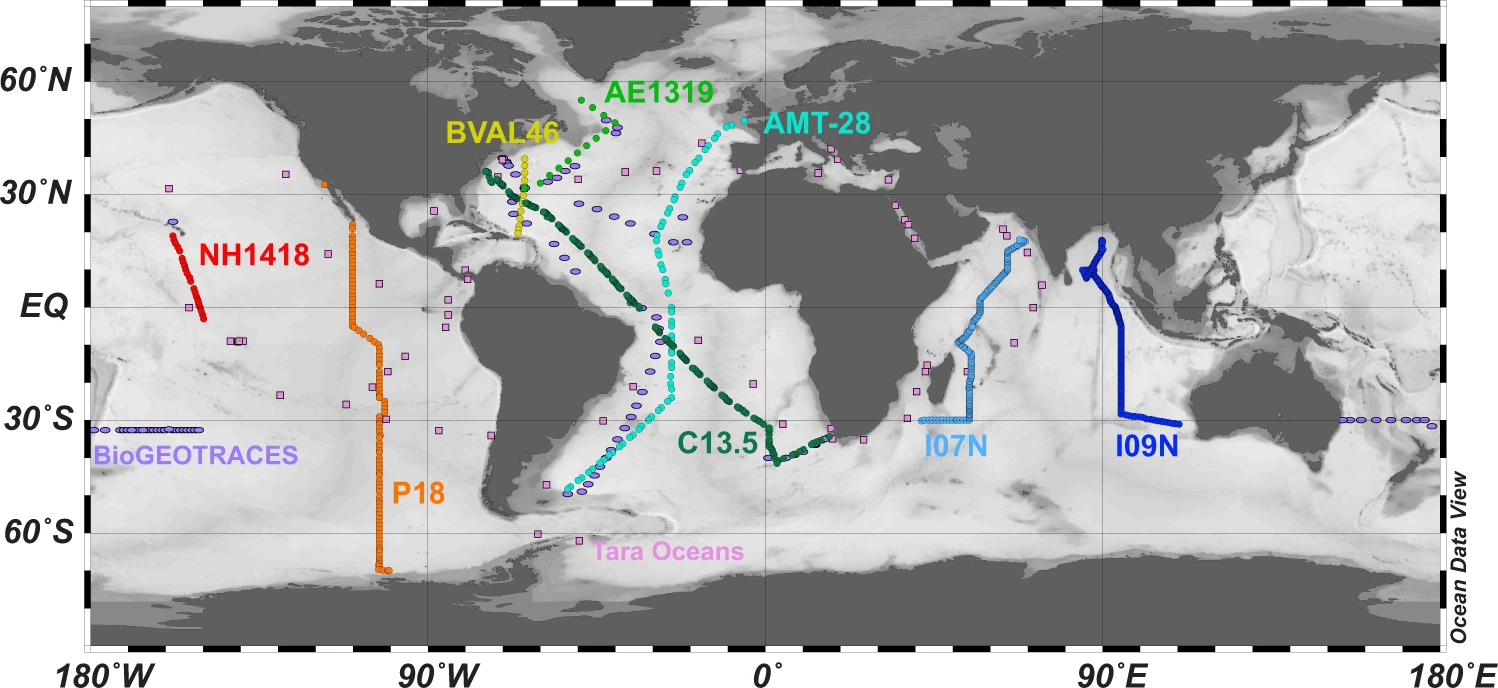
Bio-GO-SHIP
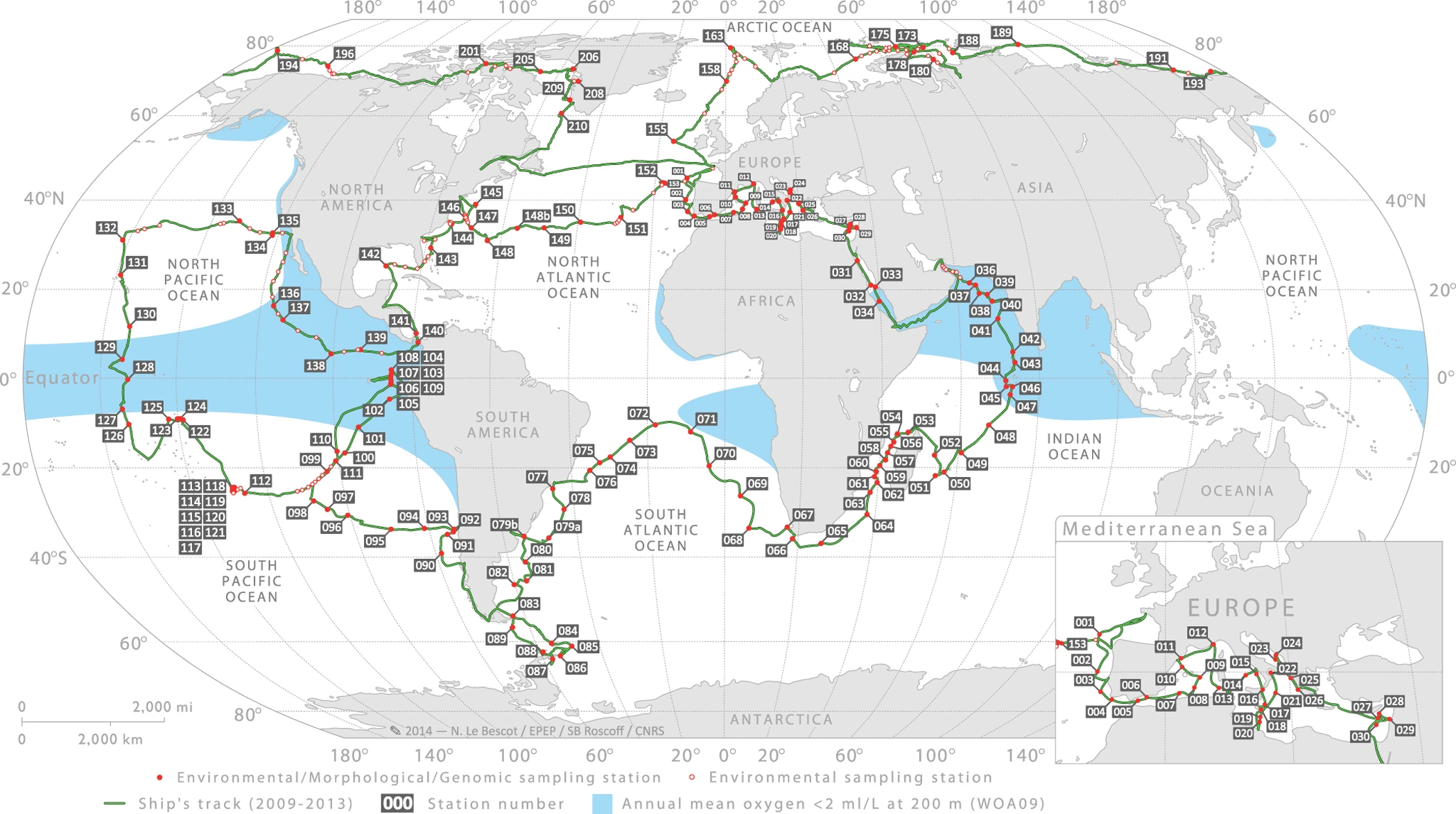
TARA Oceans Expedition
- Huge data resource, but costly ($ and compute)
- Limited depth (mostly sequence abundant things)
- Taxonomic resolution depends on mapping to a database
- Size fractionation a complication for TARA (others are > 0.2 µm)
(3) Global shotgun metagenomics = the downsides
(4) Global shotgun metabarcoding
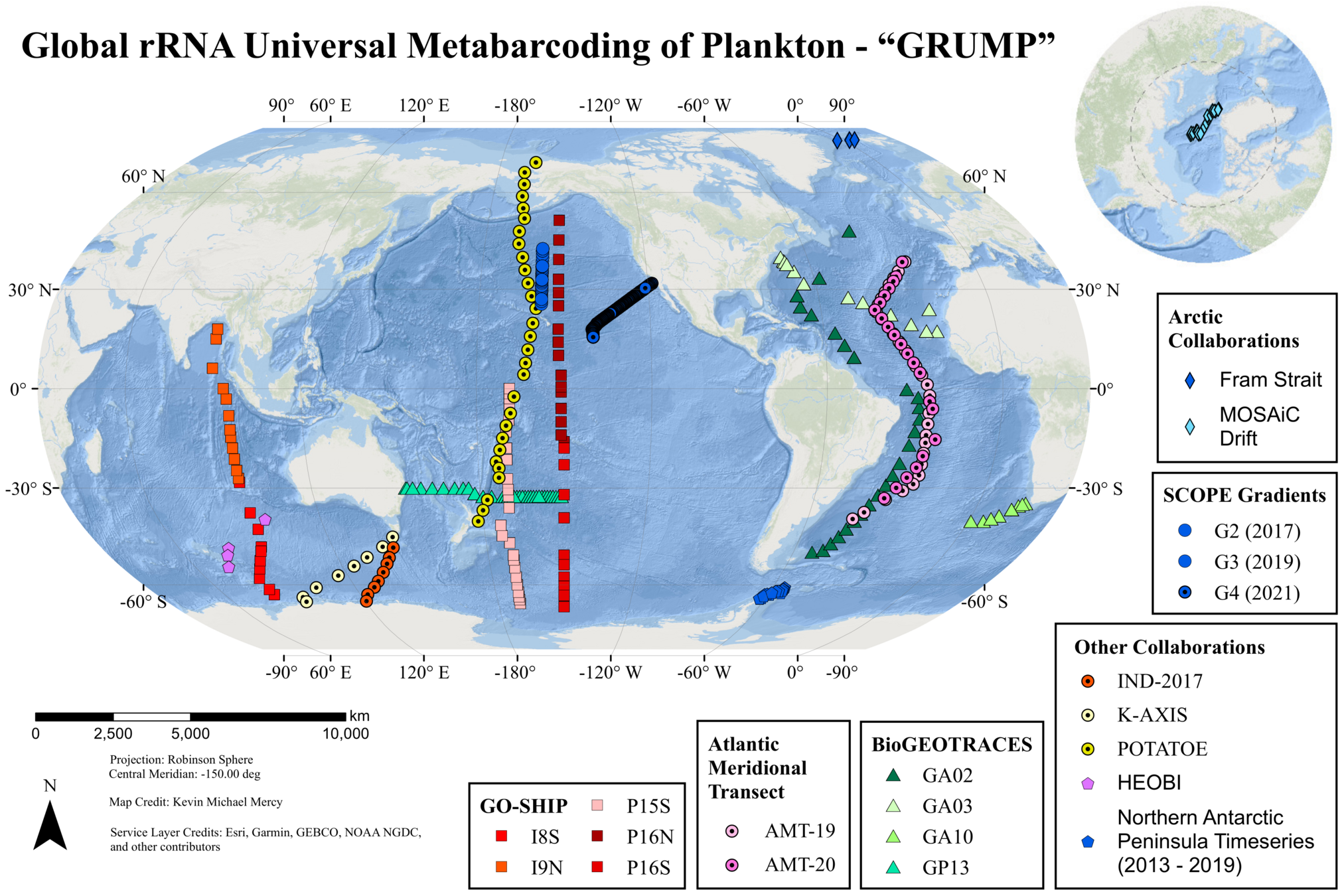
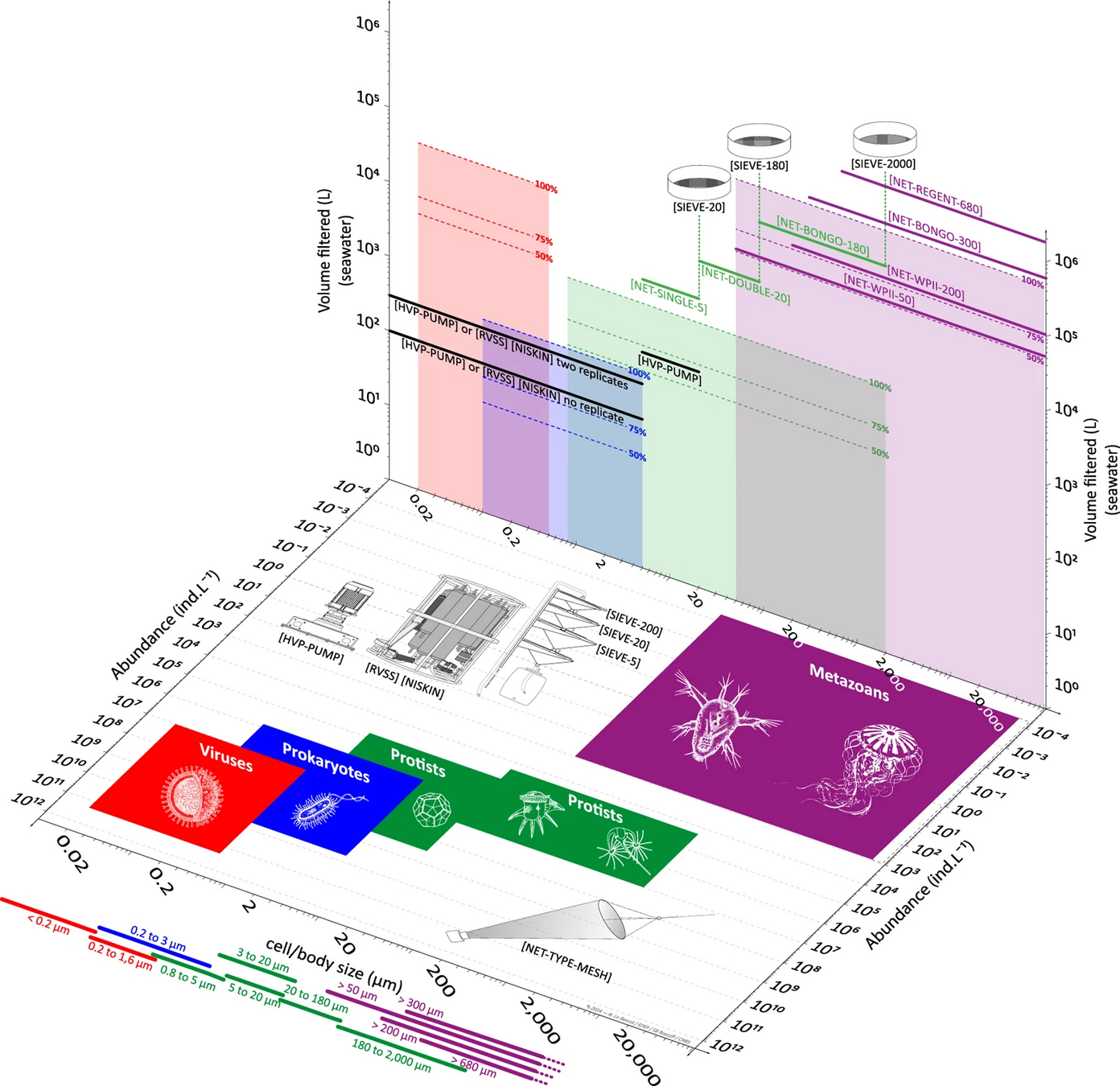
- Broad organismal range (Archaea - Zooplankton)
- Unfractionated samples (> 0.2 µm)
Universal



Different same primers, different same regions of barcode
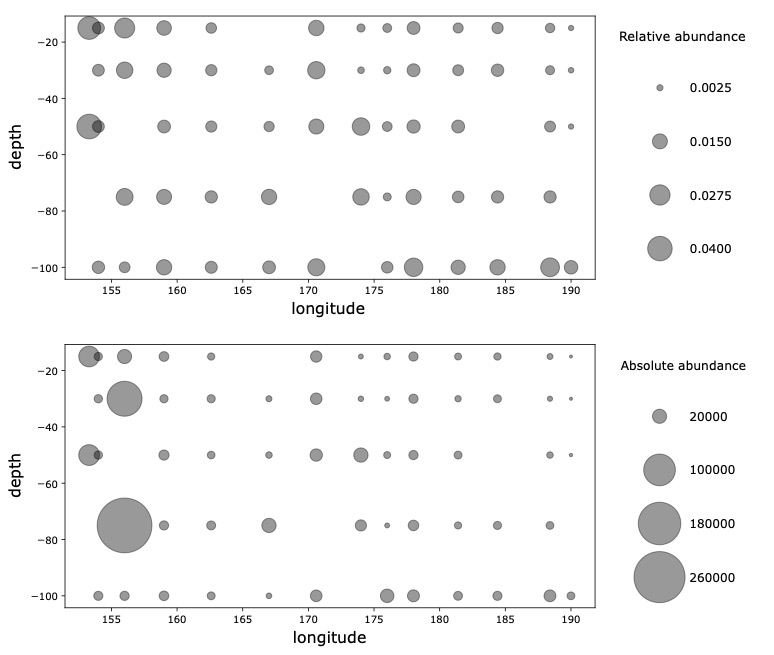
- GRUMP Rank Abundance Distributions
- Other interesting metrics
- Going beyond relative abundance
- How much trait information do we need?
- Do we need to integrate short & long-read technologies?
(5) Data & Directions
(5) GRUMP RADs (P16N/S)
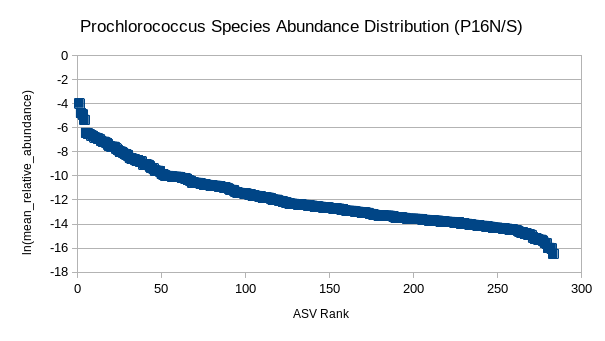
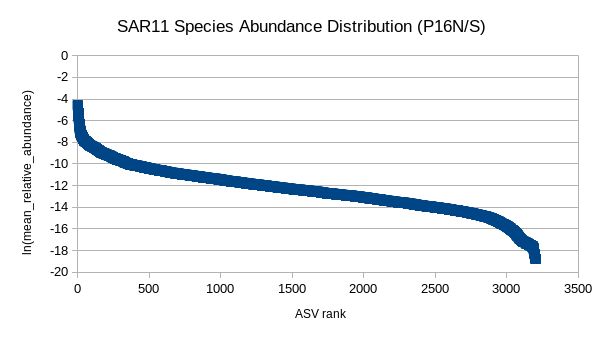
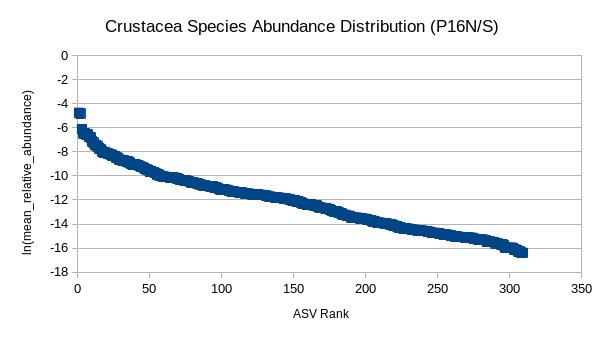
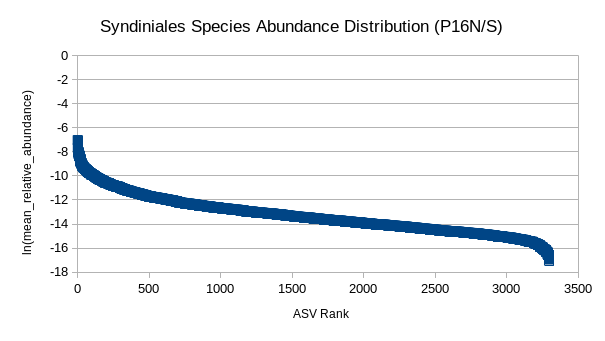
Are organism ranks stable?
How do RADs differ:
- Across depth?
- Ecological province?
- Trophic level?
(5) Other interesting metrics
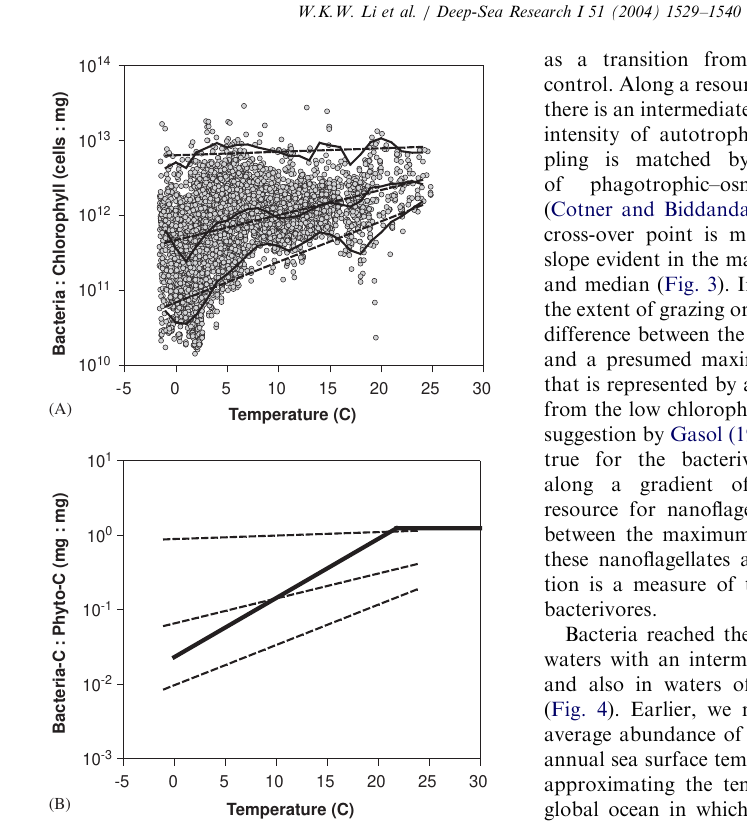

Microheterotroph: Phytoplankton ratios
Is this true in the Southern Ocean or other, unusual enviroments? What about metazoans or other taxa?
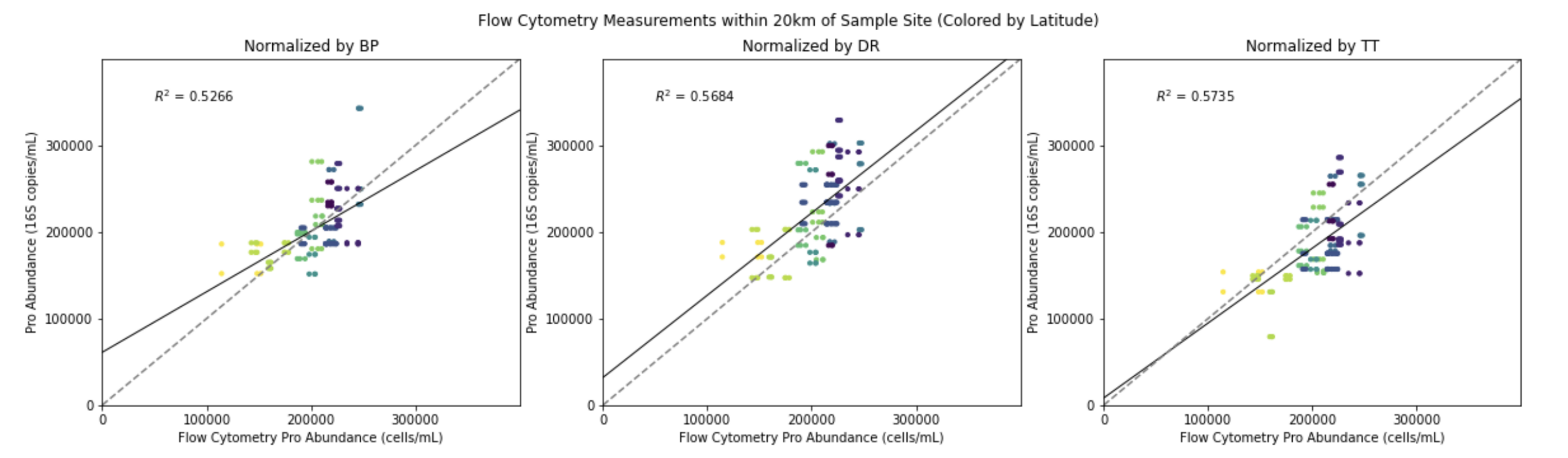
(5) Beyond relative abundance
Compositional data is not ideal. What to do?
Lexi
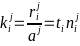
Enrico & Mick
Use paired data such as FACS as "anchor"
Analyze with internal standards (spike-in)
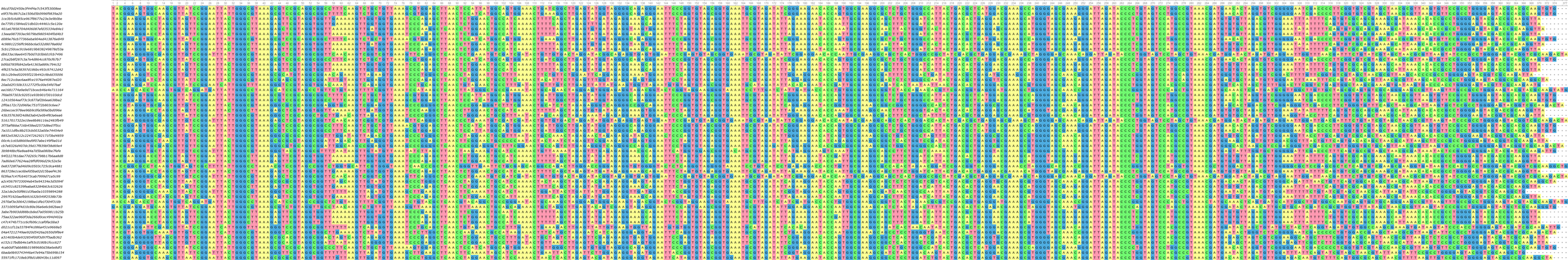
ASV DNA data

ASV phylogeny
(5) Linking ASVs to traits
How much trait information is needed to interpret macroecological patterns?
- How to robustly intercompare data from different rRNA regions?
Advantages of full-length rRNA database:
- Allows intercomparisons with legacy datasets
- Potentially improves taxonomic resolution of ASV data
Dueholm et al. (2020) mBio, e01557-20

(5) Integrating short-, long-read technologies
Environmental DNA/RNA
Long-read sequencing (e.g. PacBio CCS)
Database of full-length 16S rRNA
The End

Cost of identifying organisms
Methods summary: Pros and cons
Other metrics
- Species area relationship (SAR)
- Distance decay (Florida straits)
- Taylor's power law
Macroecology, new data sources, CBIOMES 2023
By jcmcnch
Macroecology, new data sources, CBIOMES 2023
- 57