Marine Microbial Ecology at StFX
StFX Biology department, March 31st, 2021
Jesse McNichol, PhD, Biological Oceanography
Postdoctoral Scholar, USC

The microbial tree of life
- Microbes are a "living fossil record" of biosphere evolution
- Have mapped diversity but cannot yet explain it
- What is the "logic" behind the tree's structure (traits, niches)?
- What are the ecological and environmental implications?

Ecosystem-centred research




- Goal = holistic under-standing of ecosystem
- Methods = puzzle pieces
- Whole puzzle = "sketch" of traits / niche
Proposed program
- Holistic program driven by broad vision
- Accomplished with diverse methods (range of depths / breadths)
-
Built with iterative and interconnected research projects
- Different experience levels and (inter)departmental collaborations


Methods
- Diverse techniques that represent different parts of the puzzle

PCR-based quantification of 16S & 18S SSU rRNA
"Phylogenetic stains" with DNA probes & (stable) isotope tracers
- Culture-based genomics
- Single-cell genomics
- Shotgun metagenomics
- Microoxic/anoxic cultivation
- Physiological measurements
- Analytical chemistry assays

Areas of research
- Cultivation:
- Genomics, Physiology
- Barcoding surveys
- Process studies




Cultivation


- Pure cultures are essential for interpreting environmental 'omics data
- Isolation is a great introductory project:
- Most microbes are unknown to science
- Develop based on interests / strengths
- Isolation is a great introductory project:

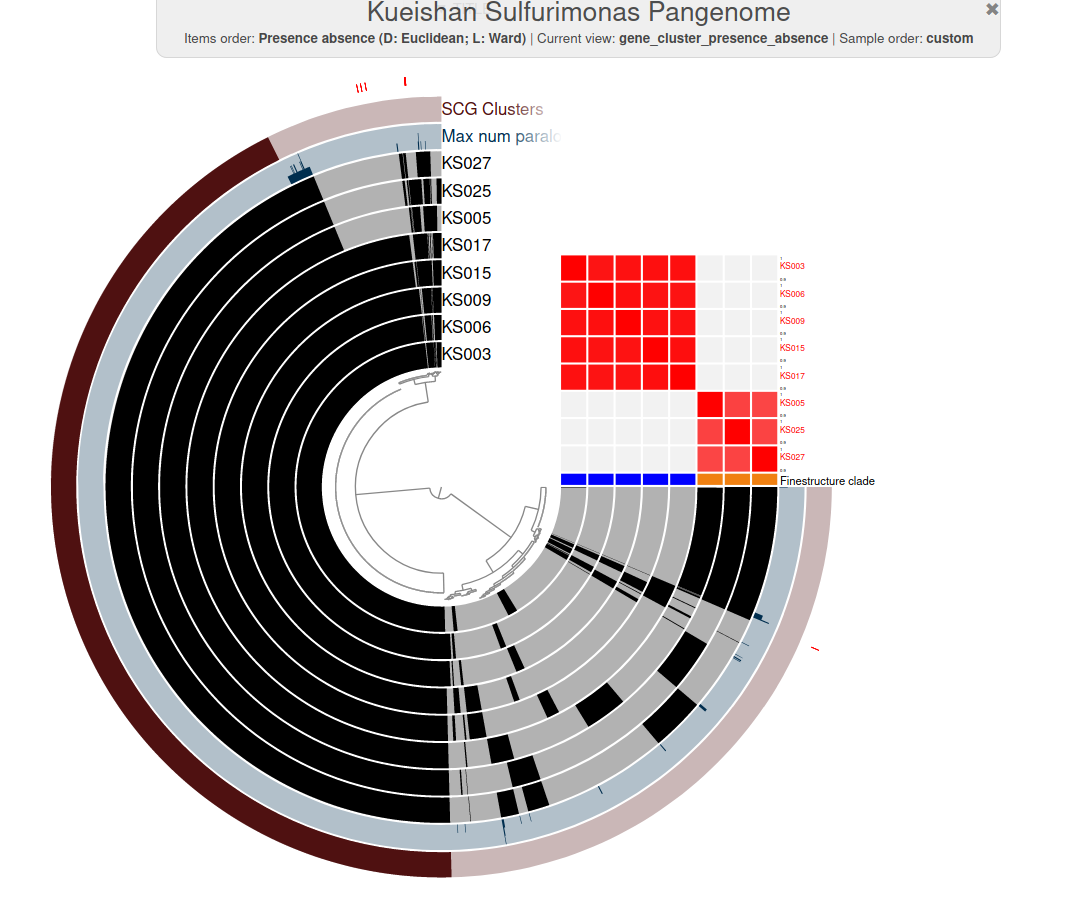
Cultivation & genomics
- Sequencing genomes of multiple isolates shows how speciation and evolution may occur in nature


Pure cultures or environmental genomes
(Pan)genomic analysis


Understanding of speciation
Plot made with anvi'o
Cultivation & physiology


- Develop and test hypotheses about biochemical basis of ecological niches

Specific hypotheses that can be used to guide future work ('omics / biochemistry)
Cultivation @ StFX
- Set up equipment for O2-sensitive microbes
- Retrieve cultures / samples from WHOI, CUHK
- Train students in isolation, cultivation
- Medium term: develop 'omics methods for cultures
- Long term: Targeted isolation campaigns, biochemistry




Barcoding surveys
- Barcoding data is used to map microbes across time and space
- 515Y / 926R primers target all cellular life






Microbe art: @claudia_traboni
Barcoding surveys
- Our work is extending observations across oceans

Barcoding surveys @ StFX
- Set up bioinformatic infrastructure
- Develop "bite-size" projects:
- Biogeographic, time-series data
- Analysis, interpretation, "data science"
- Contribute to modelling work
- Med. term: Build lab infrastructure (eDNA)
- Long term: Apply to local "model systems"




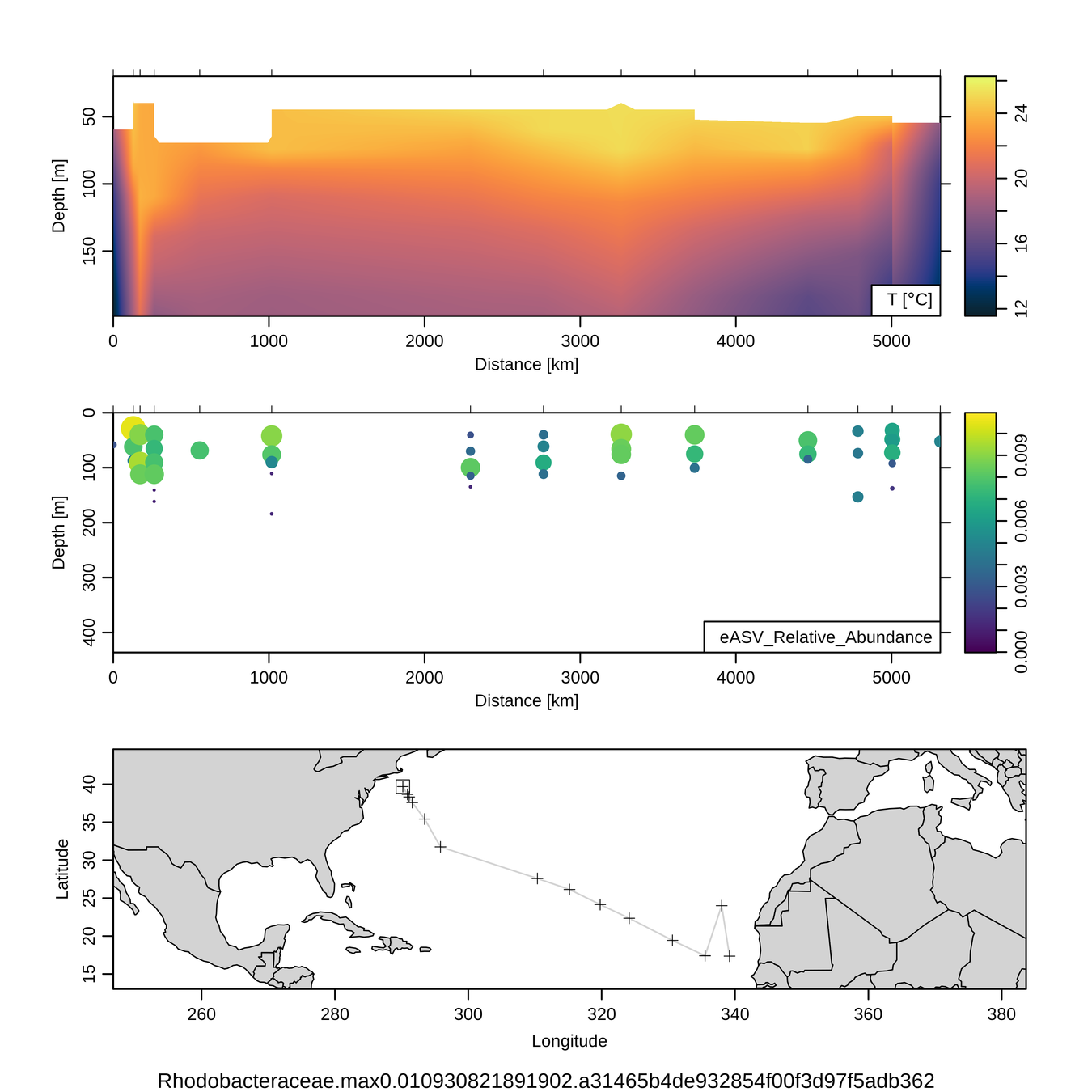
Process studies (hydrothermal vents)


- How much CO2 do hydrothermal vent microbes fix?
- What factors influence their growth?



- Barcoding showed major shift due to oxygen
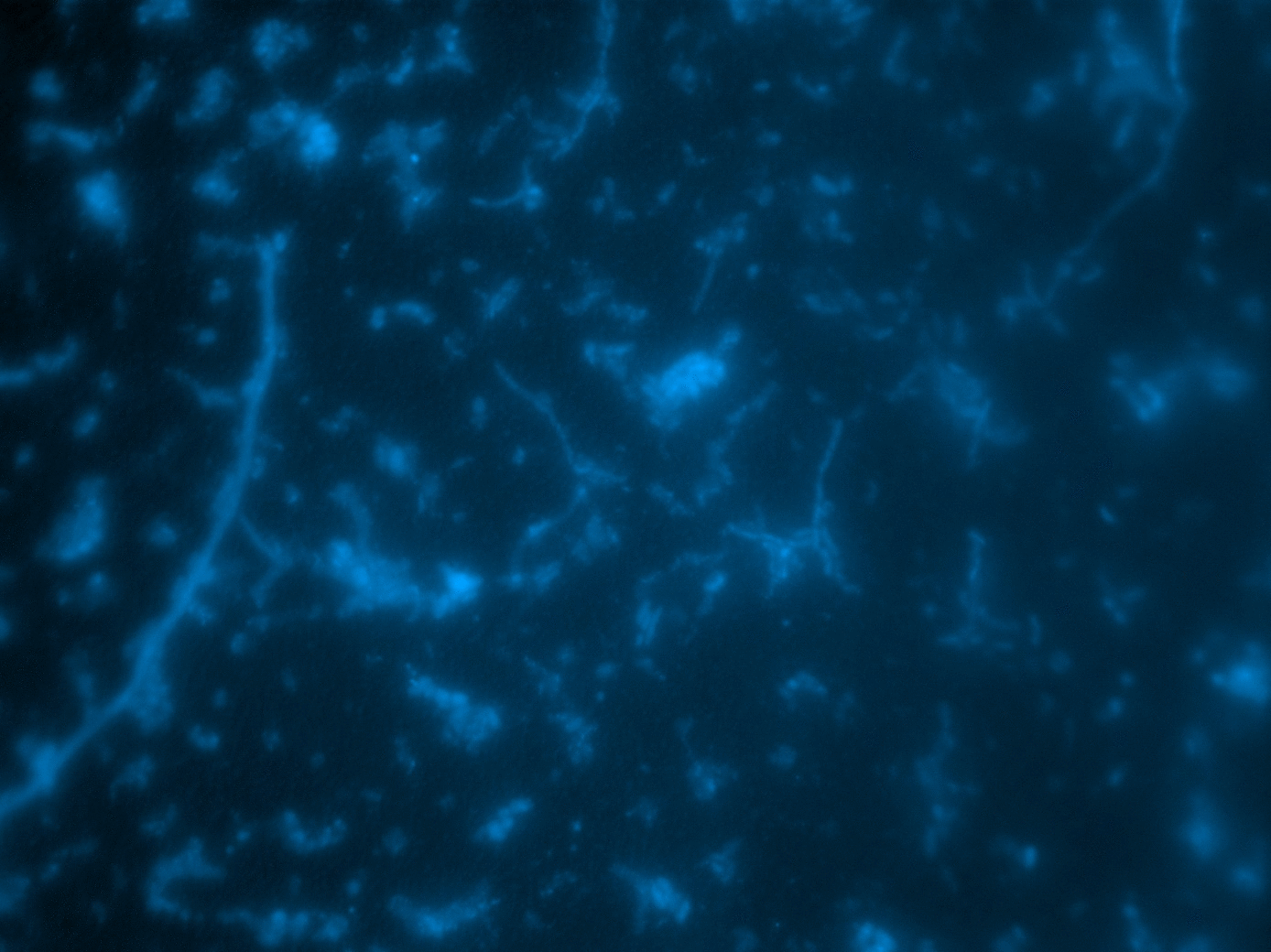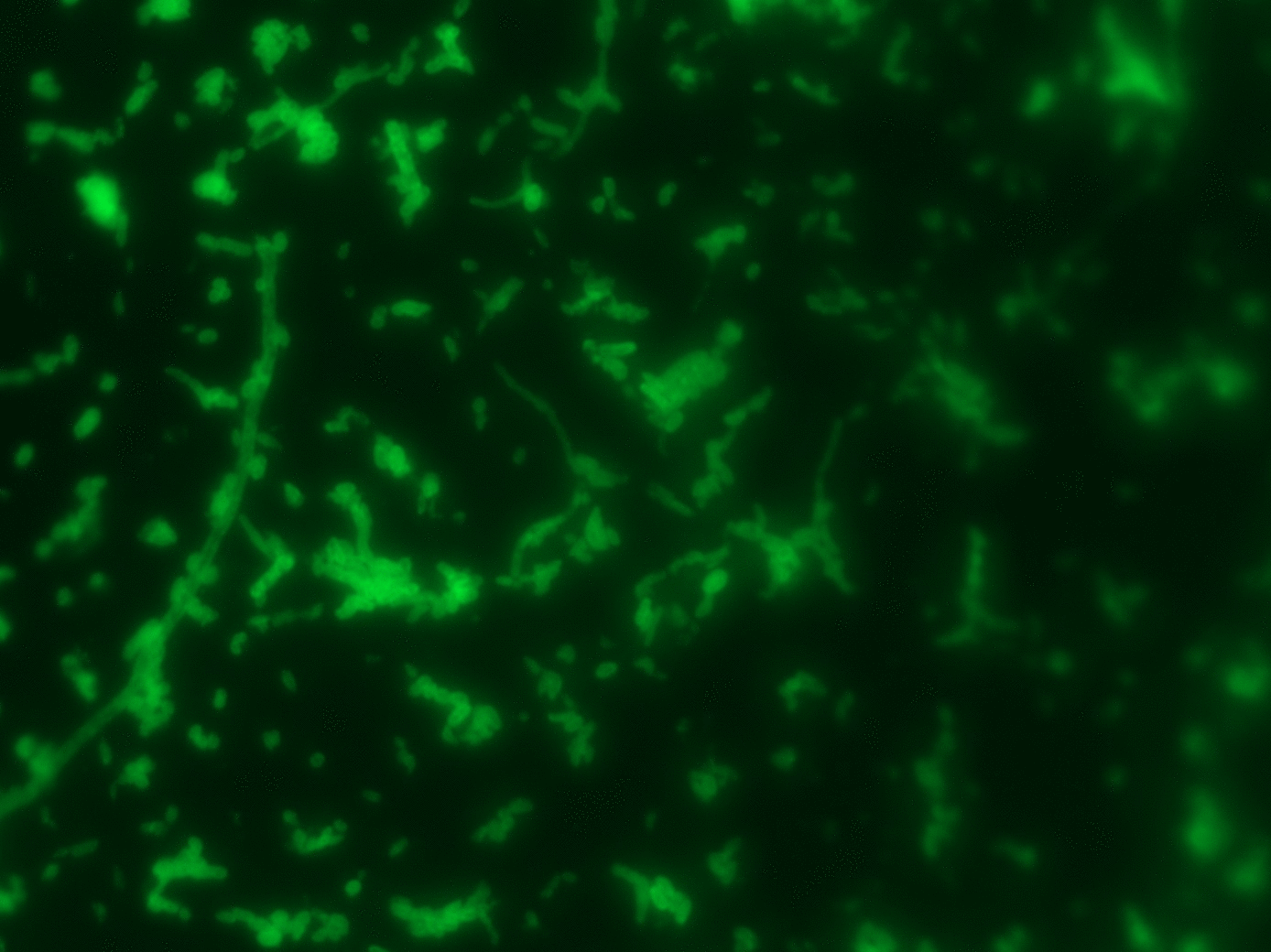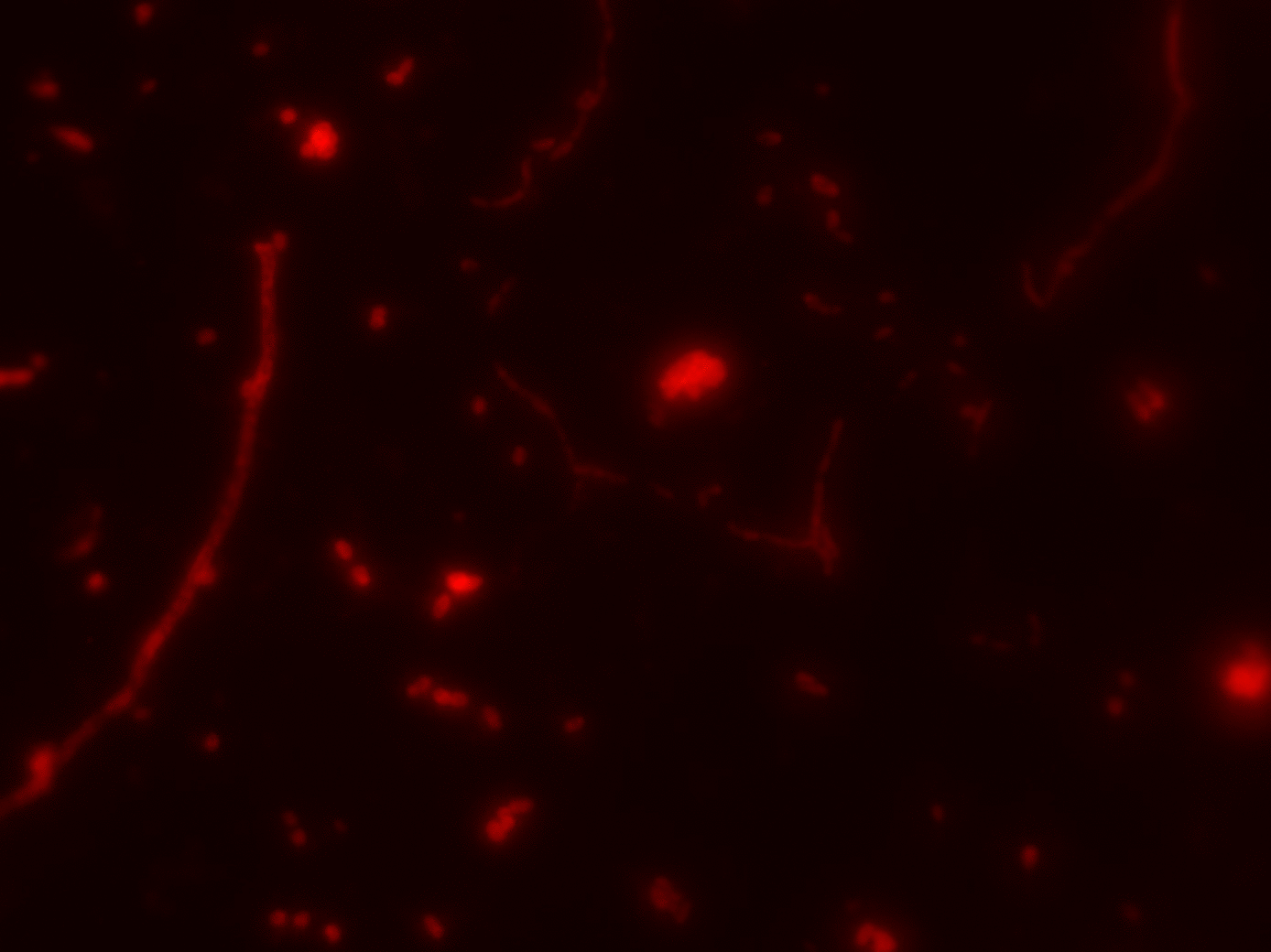
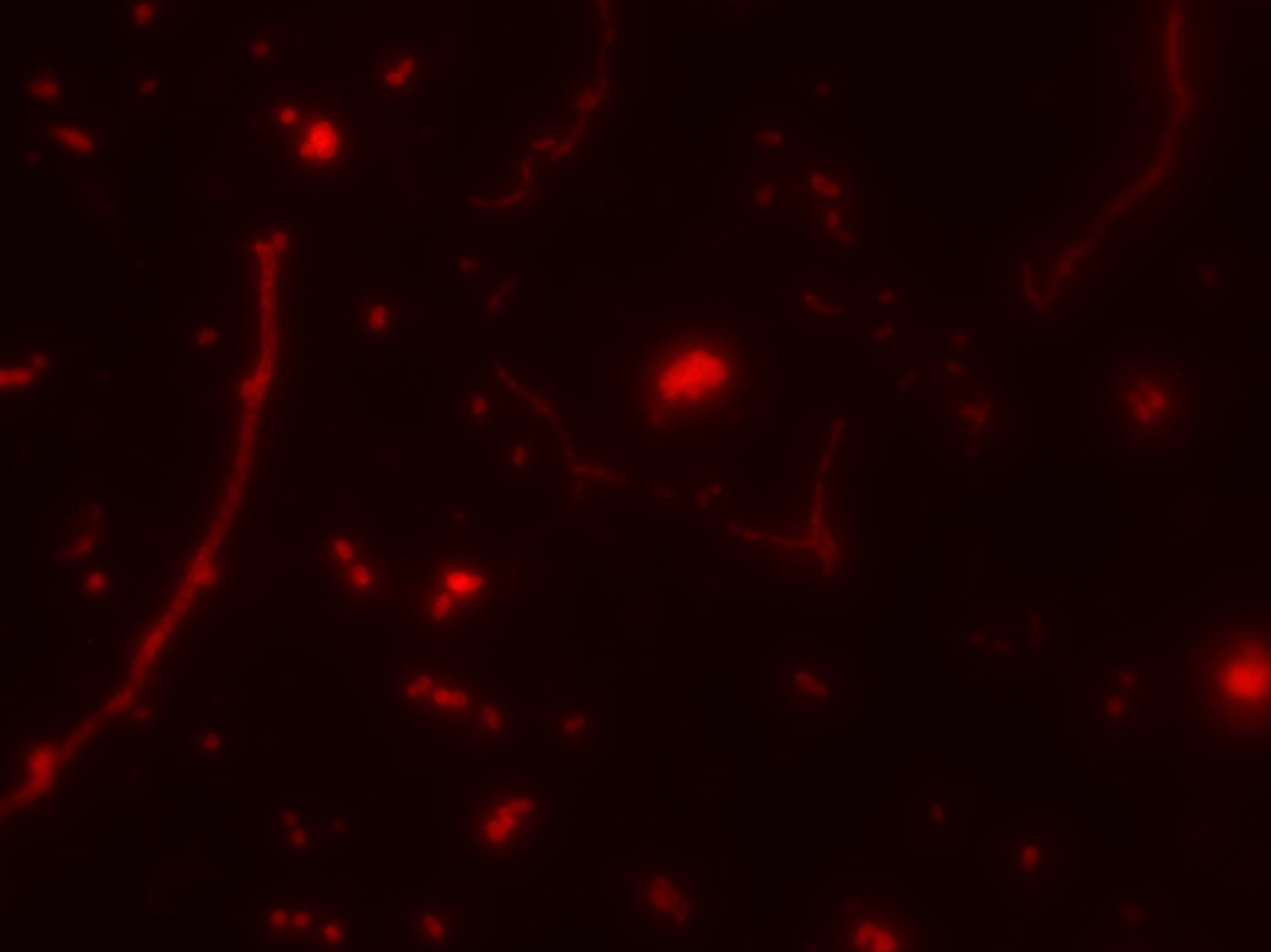
- FISH + SIP quantified CO2 fixation at single-cell level (NanoSIMS)


Oxygen caused major shift in community structure over ~ 24 h
FISH
SIP
Process studies (hydrothermal vents)
- Set up lab infrastructure for FISH
-
Medium term:
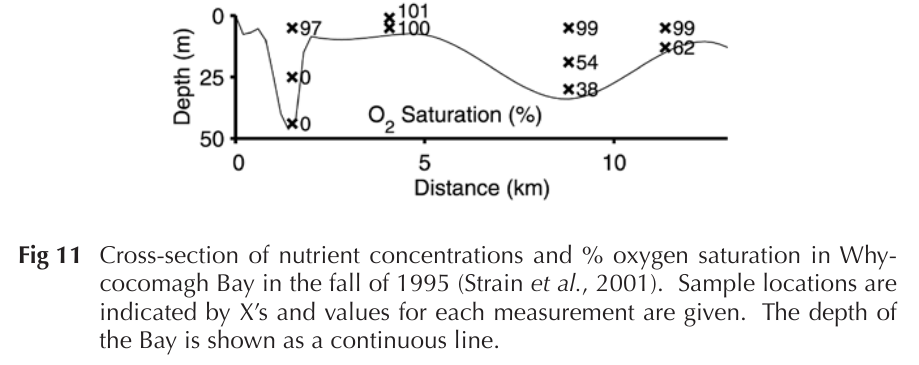
- Investigate Whycocomagh bay as model system:
- Water anoxic at 10 - 15 m, contains sulfide
- These conditions favour chemoautotrophs
- (Inter)departmental collaborations (microbiomes?)
- Investigate Whycocomagh bay as model system:

Process studies @ StFX

- Methods inform and interact with one another
- Points to "microbial natural history"

Holistic picture







Native Microbiota CURE
- Based on undergraduate botany experience (Native Flora)
- Students collect microbial "herbaria":
- Characterize with barcoding
- Visualize with FISH
- Attempt to cultivate
- Goal: Develop "microbial natural history"
- Bring new scientists into the fold
- EDI objectives
- X-Oceans Outreach
- Discover new taxa, study sites
- Bring new scientists into the fold


Summary
- Samples / data:
- Global barcoding data
- Vent strains / samples
- (meta)genomic data
- Wet lab skills:
- HTS* library prep
- Low O2 cultivation
- FISH (& probe design)
- Analytical chemistry
- Bioinformatics skills:
- Barcoding (qiime2)
- 'omics (anvi'o)
- R/python/bash, snakemake**
*HTS=High throughput sequencing
**tinyurl.com/mgprimereval



Why microbial ecology at StFX?
- Opportunity to focus on fundamental biological research
- Build model (eco)systems, interdepartment collaborations
- Diverse questions / techniques fits with liberal arts education
- Flexible, student-centred projects; CURE
- Maritimes offers a wealth of natural marine environments
- Collaborations in oceanography, ecosystem monitoring



Acknowledgements
WHOI: Stefan Sievert, Jeff Seewald, Niculina Musat, Craig Taylor
CUHK/Academia Sinica: Haiwei Luo, Annie Wing-Yi Lo, Benny Chan
USC: Jed Fuhrman, Yi-Chun Yeh, Bruce Yanpui Chan, Paul Berube, Steven Biller, Mick Follows, Enrico Ser-Giacomi








Works cited (ideas and images)
| Slide | Citation |
|---|---|
| 1 (tree image) | A. Spang, T. J. G. Ettema, Microbial diversity: The tree of life comes of age. Nat Microbiol 1, 16056 (2016). |
| 2 (ocean diagram) | M. Hügler, S. M. Sievert, Beyond the Calvin Cycle: Autotrophic Carbon Fixation in the Ocean. Annu. Rev. Marine. Sci. 3, 261–289 (2011). |
| 8 (speciation diagram) | R. Stepanauskas, et al., Gene exchange networks define species-like units in marine prokaryotes. bioRxiv, 2020.09.10.291518 (2020). |
| 9 (protein image) | J. McNichol, S. M. Sievert, Reconciling a Model of Core Metabolism with Growth Yield Predicts Biochemical Mechanisms and Efficiency for a Versatile Chemoautotroph. bioRxiv, 717884 (2019). |
| 10 (abundance plot) | J. A. Fuhrman, J. A. Cram, D. M. Needham, Marine microbial community dynamics and their ecological interpretation. Nature Reviews Microbiology 13, 133–146 (2015). |
| 13 (incubation plot) | J. McNichol, et al., Assessing microbial processes in deep-sea hydrothermal systems by incubation at in situ temperature and pressure. Deep-Sea Res. Pt. I 115, 221–232 (2016). |
| 12 (FISH + SIP diagram) | J. McNichol, et al., Primary productivity below the seafloor at deep-sea hot springs. PNAS 115, 6756–6761 (2018). |
| 15 (Water column diagram) | P. M. Strain, P. A. Yeats, The Chemical Oceanography of the Bras D’Or Lakes. Proc NSIS 42, 37–64 (2002). |
| 17 (book image) | B. D. Dyer, A Field Guide to Bacteria, Illustrated edition (Comstock Publishing Associates, 2003). |
Unless otherwise noted, images are either my own work, ⓒWHOI, or from wikimedia commons
Understand to protect

- Microbes are the "wires" connecting global chemical cycles
- Climate, nutrient cycling, habitability
A microbial ecology lab at StFX
- Ultimate goal:
- Niche / traits of marine bacteria
- Long-term, iterative program:
- Undergrads, grad students, postdocs
- (Inter)departmental collaborations
- Bring materials, expertise, collaborations:
- Quickly begin research with 'omics / barcoding data
- Samples (e.g. for cultivation)
- Longer term goals once established:
- Model ecosystem (Whycocomagh bay?)
- Native Microbiota CURE
- Monitoring collaborations


My experience
- B.Sc., Biology, Mount Allison (2003 - 2008)
- Government researcher (2008-2011)
- Environment Canada
- NRC (Algal Biofuels)
- PhD in Biological Oceanography, MIT/WHOI (2011-2016)
- Postdocs:
- CUHK (2017)
- USC (2018 - present)



Why study microbial ecology?
- Like astronomy, it gives us a sense of perspective...
- ...and helps us look back in time to how life evolved


Woese's tree

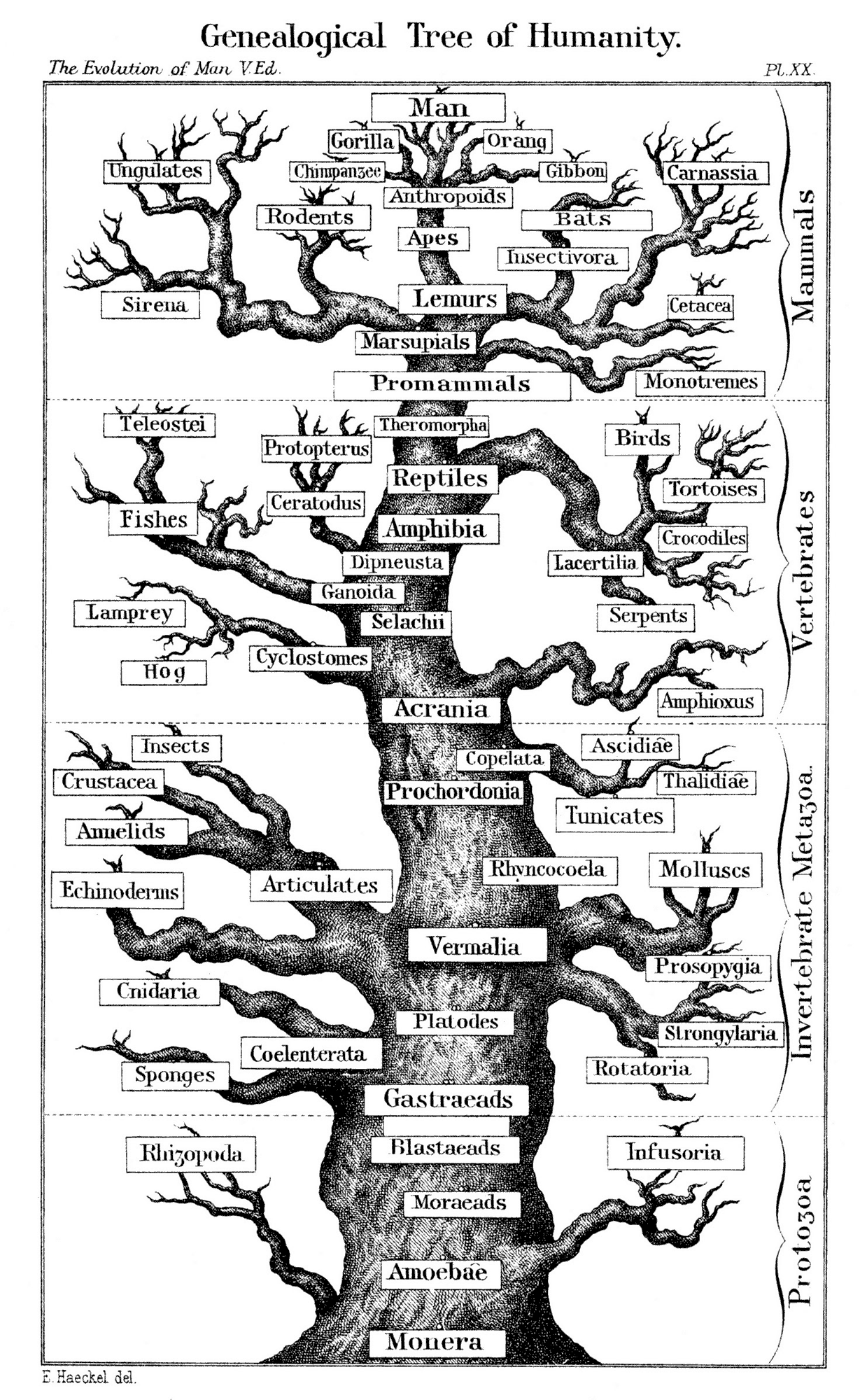
Haeckel's tree
Undergraduate research at StFX
- Microbes are everywhere, yet poorly described:
- Cultivation / surveys
- Native Microbiota
- Huge amount of 'omics data:
- "Data science"
- Tools have yet to be perfected:
- Methods development




StFX Research Mini-Lecture, March 2021
By jcmcnch
StFX Research Mini-Lecture, March 2021
An introduction to my PhD and postdoctoral research on marine microbial ecology, and how I envision a research program developing at StFX with active participation from undergraduate, graduate, and postdoctoral researchers.
- 52



