Python in Life Sciences
@juliocesar_io
https://juliocesar.io
Federating Data for Science
- About federation
- Architecture
- Tech Stack
- Ariadne GraphQL
- Apollo Gateway
- Nvidia Clara
- Hands-on demo
Agenda
About Federation
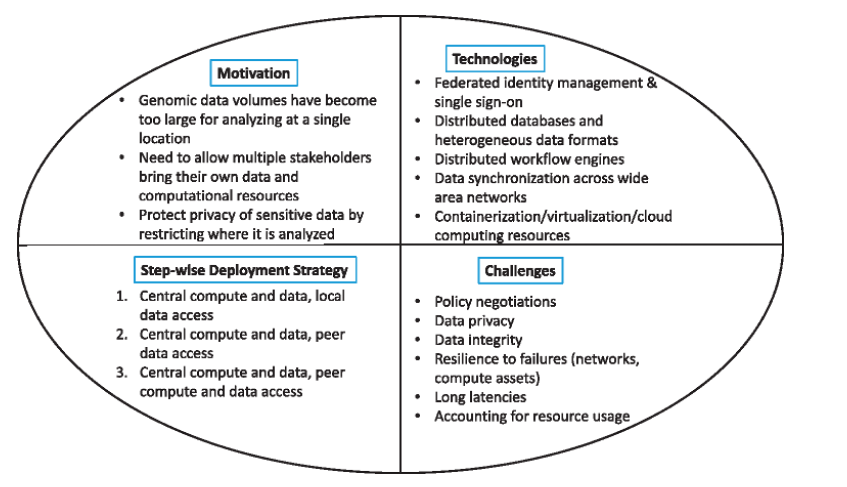
It's been a challenge in bioinformatics.
Federation is the ability of multiple independent resources to act like a single resource. Cloud computing itself is a federation of resources, so the many assets, identities, configurations and other details of a cloud computing solution must be federated to make cloud computing practical’.

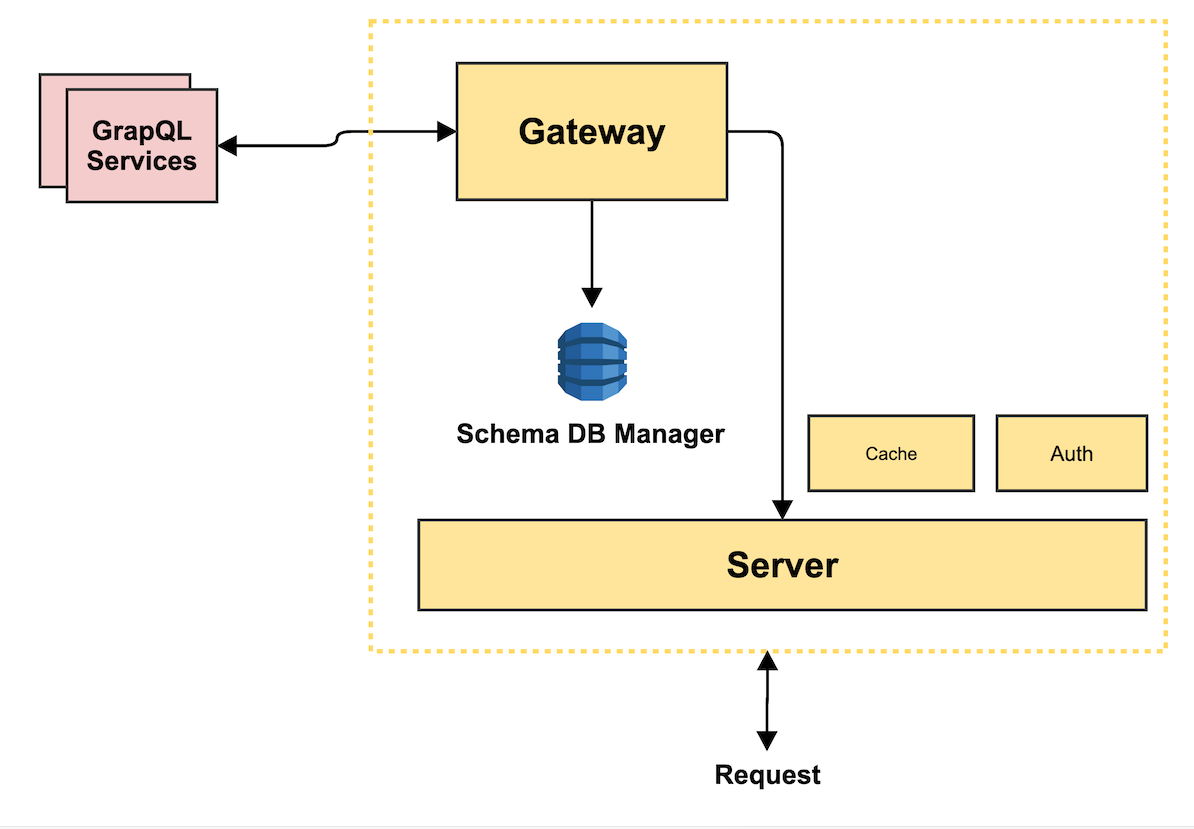
Architecture
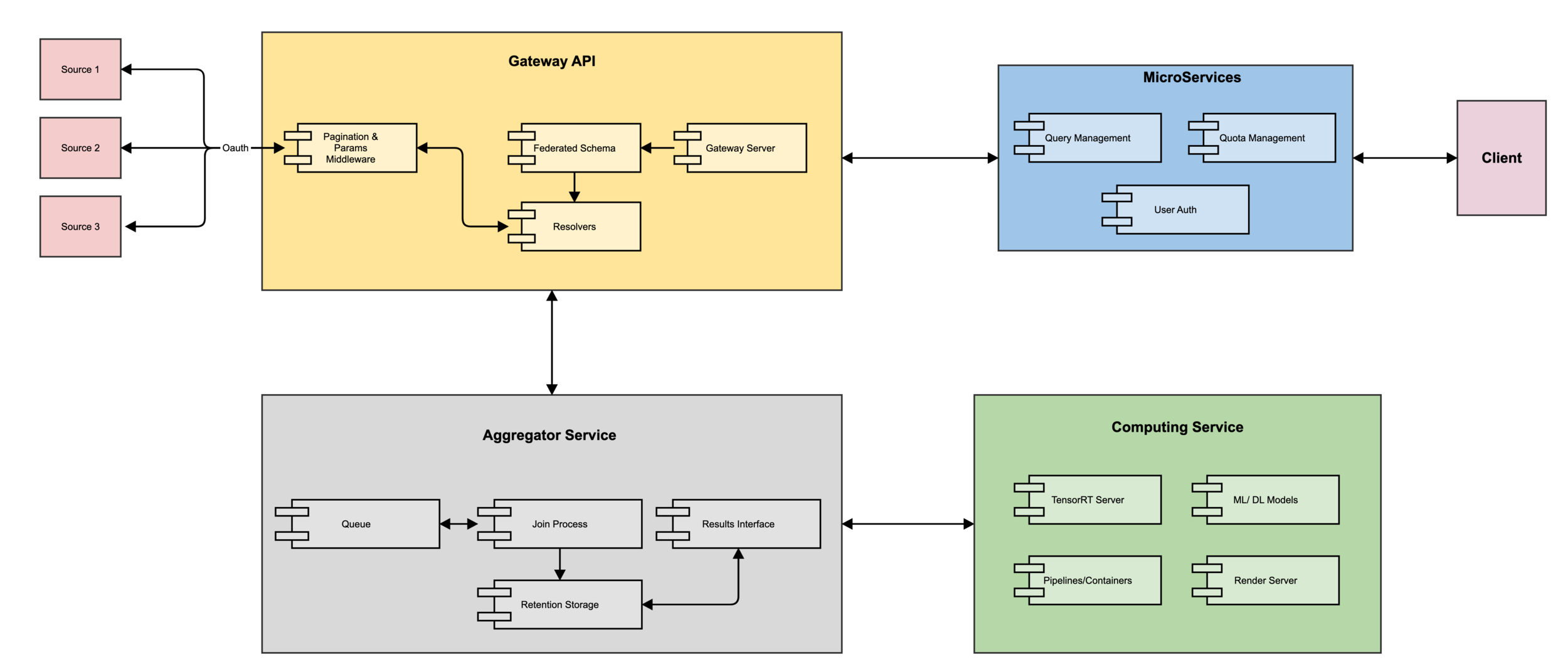
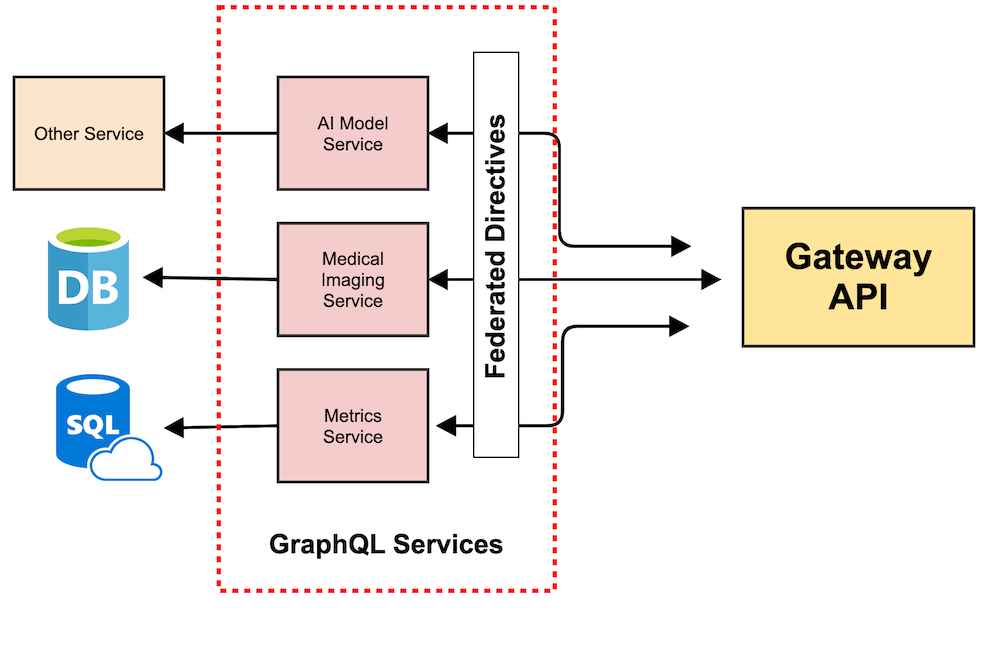
High Level Components
The Stack


-
Gateway API
- Implementing Services Ariadne/GraphQL
- Federated Server Apollo
- DynamoDB Managed Schemas
-
Aggregation Service
- HDFS
- Async tasks
- Merge results
-
Computing Service
- Nvidia Clara SDK (Models, TensorRT Server, DICOM/PACS Interface)
- Kubernetes / Docker / Helm
- Management API
- Nvidia Render Server




Implementing Services
Ariadne GraphQL

- Schema First SDL
- Federated Schema
- Apollo Support
- The Python you know and love :)
Services Definition
Ariadne GraphQL

The Project structure
Ariadne GraphQL

type Query {
models(id: ID, name: String, platform: String): [Model]
}
type Model @key(fields: "id"){
id: ID!
name: String!
platform: String!
data_type: String!
kind: String!
url: String!
version: String!
}Schema Definition
Ariadne GraphQL
type Query {
medicalImage(id: ID): [MedicalImage]
}
type Model @key(fields: "id") @extends {
id: ID! @external
medicalImages: [MedicalImage]
}
type MedicalImage @key(fields: "id") {
id: ID!
type: String
format: String
url: String!
model_id: Int!
}AI Model Service
Medical Imaging Service
with the @extends and @external we reference the relationship with the AI Model Service
model = FederatedObjectType("Model")
query = QueryType()
@model.reference_resolver
def resolve_model_reference(_, _info, representation):
return get_model_by_id(representation.get("id"))
schema = make_federated_schema("schema_file.graphql", [query, model])
Resolvers Reference
Ariadne GraphQL
AI Model Service
Ariadne GraphQL
query = QueryType()
model = FederatedObjectType("Model")
medicalImage = FederatedObjectType("MedicalImage")
@model.resolve_reference
def resolve_model(representation):
model_id = representation.get('id')
return Model(id=model_id)
@model.field("medicalImages")
def resolve_model_images(obj, info):
kwargs = {
"model_id": obj.id
}
return self.ds.get_medical_image(**kwargs)
schema = make_federated_schema("schema_file.graphql", [query, model, medicalImage])Medical Imaging Service
Resolvers Reference
Making Queries on each service
Ariadne GraphQL
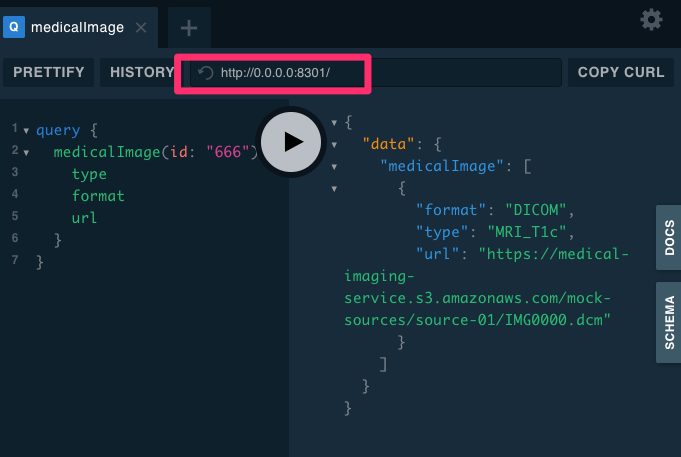
Federated Gateway on Apollo
Apollo Server
Single data graph that provides a unified interface for querying all of your backing data sources. This allows clients to fetch data from any number of sources simultaneously, without needing to know which data comes from which source.
Federated Gateway on Apollo
Apollo Server
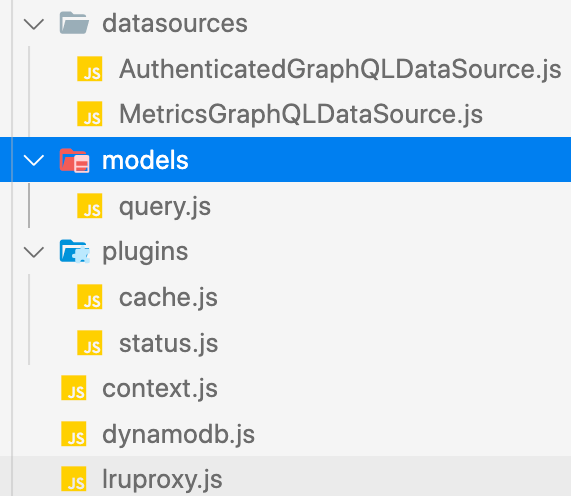
Project Structure
Apollo Server
Registering Services
Apollo Server
const gateway = new ApolloGateway({
serviceList: [
{ name: 'ai_model_service', url: 'http://localhost:4001' },
{ name: 'medical_imaging_service', url: 'http://localhost:4002' },
{ name: 'metrics_service', url: 'http://localhost:4003' }
]
});
const server = new ApolloServer({ gateway });
server.listen();Schemas and resolvers live in the implementing services. The gateway serves only to plan and execute GraphQL operations across those implementing services
A managed way to register services with DynamoDB
Apollo Server
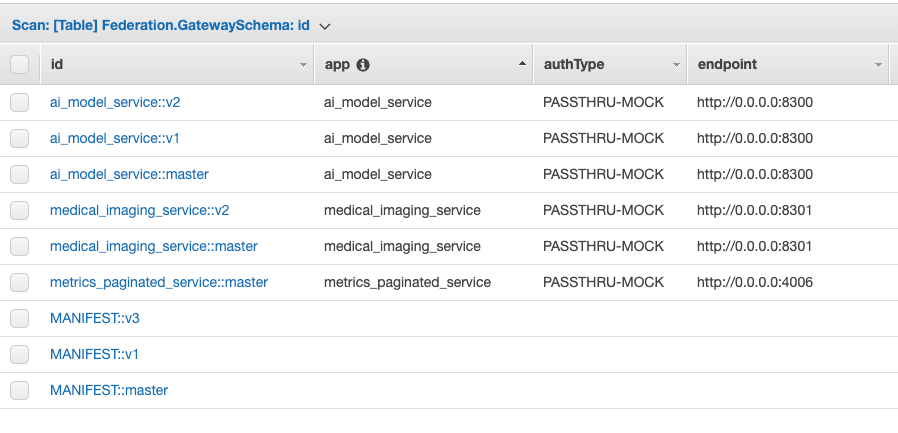
A managed way to register services with DynamoDB
Apollo Server
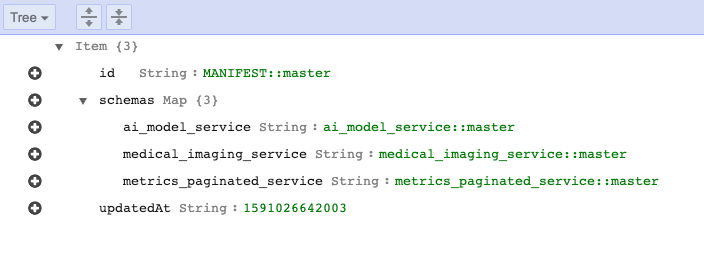
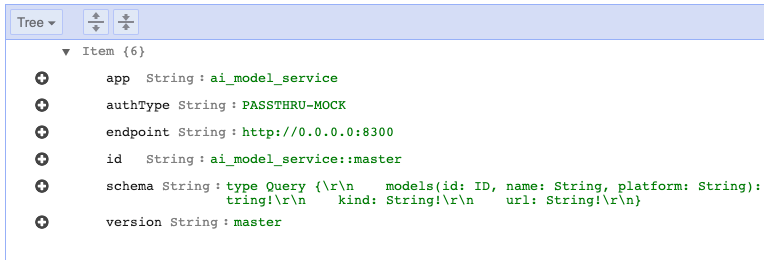
Making Federated Queries
Apollo Server

The Response
Apollo Server
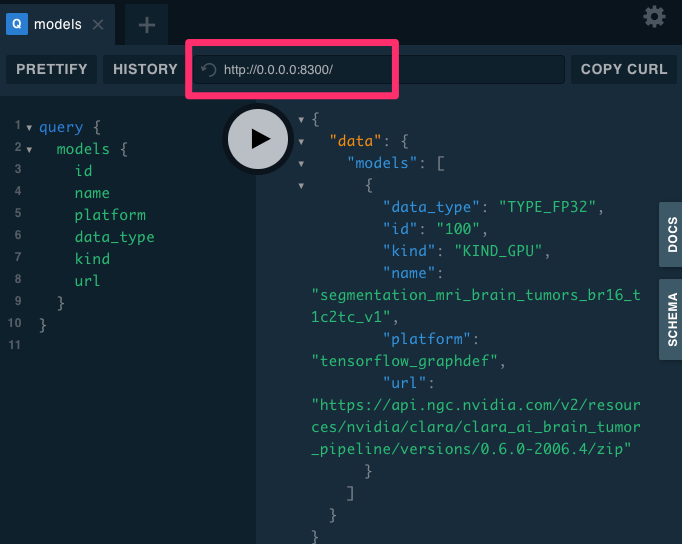
{
"data": {
"models": [
{
"id": "100",
"name": "segmentation_mri_brain_tumors_br16_t1c2tc_v1",
"platform": "tensorflow_graphdef",
"data_type": "TYPE_FP32",
"kind": "KIND_GPU",
"url": "https://api.ngc.nvidia.com/v2/resources/nvidia/clara/clara_ai_brain_tumor_pipeline/versions/0.6.0-2006.4/zip",
"medicalImages": [
{
"id": "666",
"type": "MRI_T1c",
"format": "DICOM",
"url": "https://medical-imaging-service.s3.amazonaws.com/mock-sources/source-01/IMG0000.dcm"
},
{
"id": "667",
"type": "MRI_T1c",
"format": "DICOM",
"url": "https://medical-imaging-service.s3.amazonaws.com/mock-sources/source-01/IMG0001.dcm"
},
{
"id": "668",
"type": "MRI_T1c",
"format": "DICOM",
"url": "https://medical-imaging-service.s3.amazonaws.com/mock-sources/source-01/IMG0002.dcm"
}
.....
...
..
.Computing Service
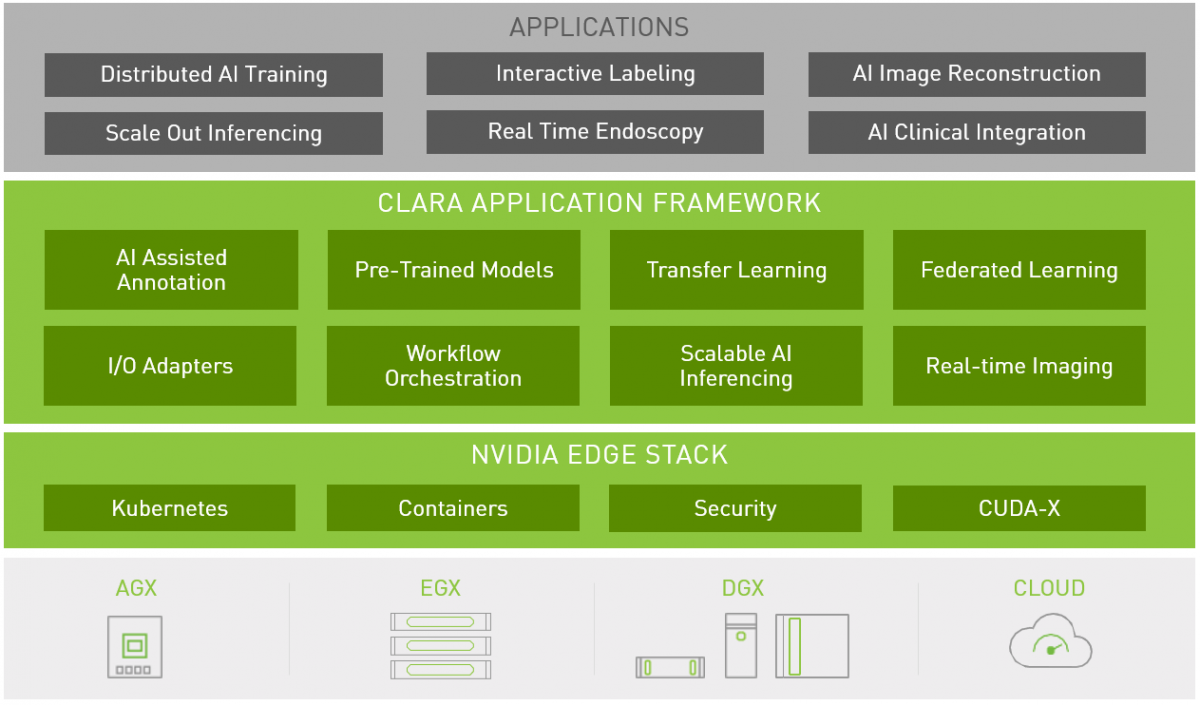
Nvidia Clara
Managing and scaling Imaging, Genomics, and Video Processing workloads. It uses Kubernetes under the hood to define a multi-staged container-based pipeline.
Architecture
Nvidia Clara
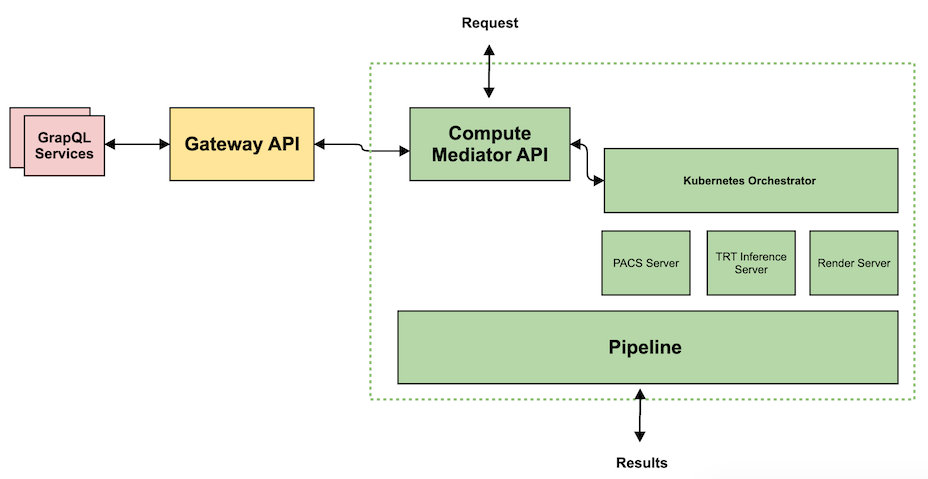
Service Interface Definition
Nvidia Clara
Project Deployment in AWS
Nvidia Clara
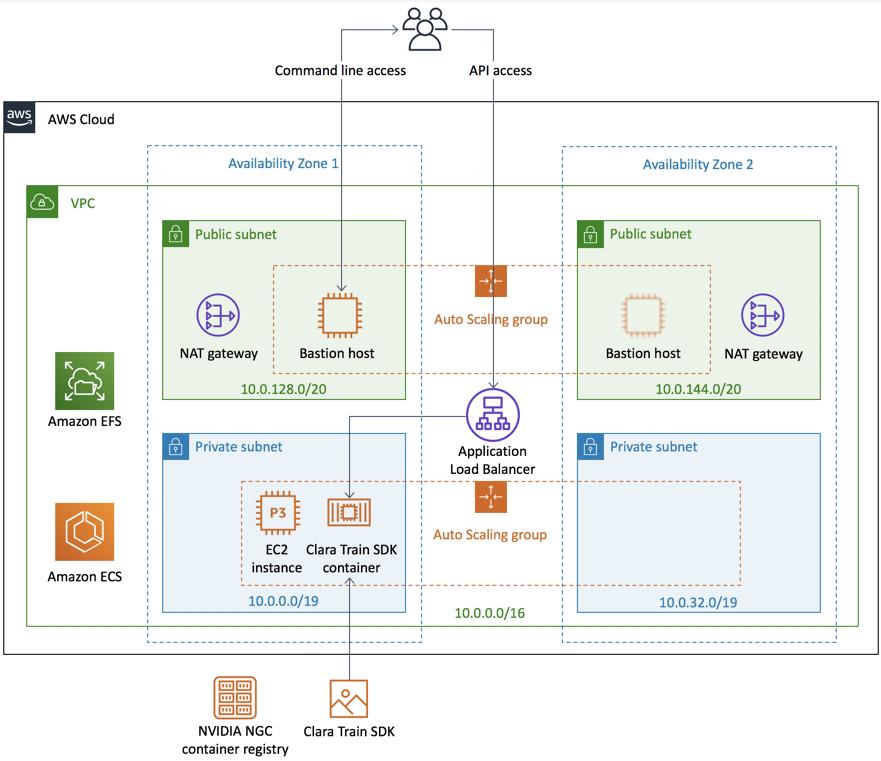
Using the services to run a model with Federated data
Nvidia Clara
{
"pipeline_tag": "brain-tumor-pipeline",
"input_tag": "dcm",
"use_cache": true,
"query": "query {\r\n models(name:\"segmentation_mri_brain_tumors_br16_t1c2tc_v1\"){\r\n id\r\n name\r\n platform\r\n data_type\r\n kind\r\n url\r\n medicalImages{\r\n id\r\n type\r\n format\r\n url\r\n }\r\n }\r\n}\r\n"
}{
"status": "RUNNIG",
"jobs": "http://ec2-18-220-78-85.us-east-2.compute.amazonaws.com:32002/jobs/",
"render": "http://ec2-18-220-78-85.us-east-2.compute.amazonaws.com:8080/renderserver"
}Response
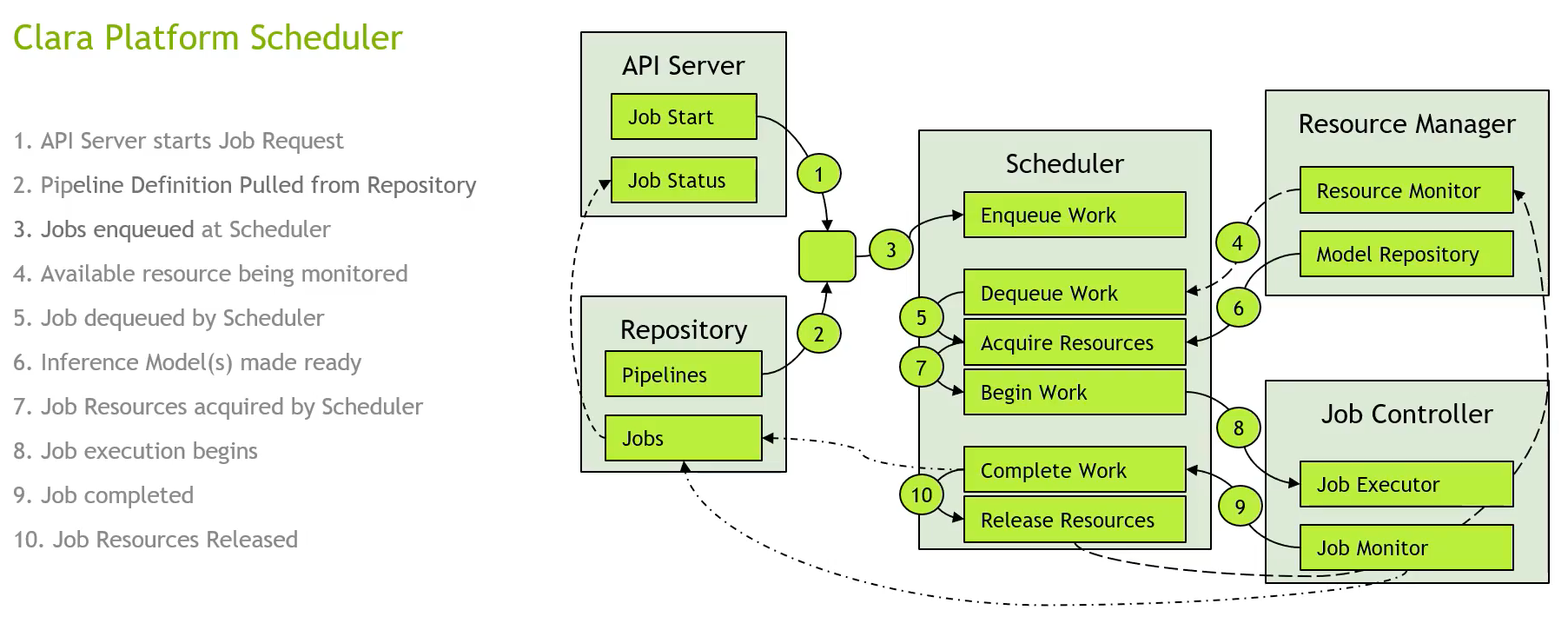
The Clara Process
Nvidia Clara
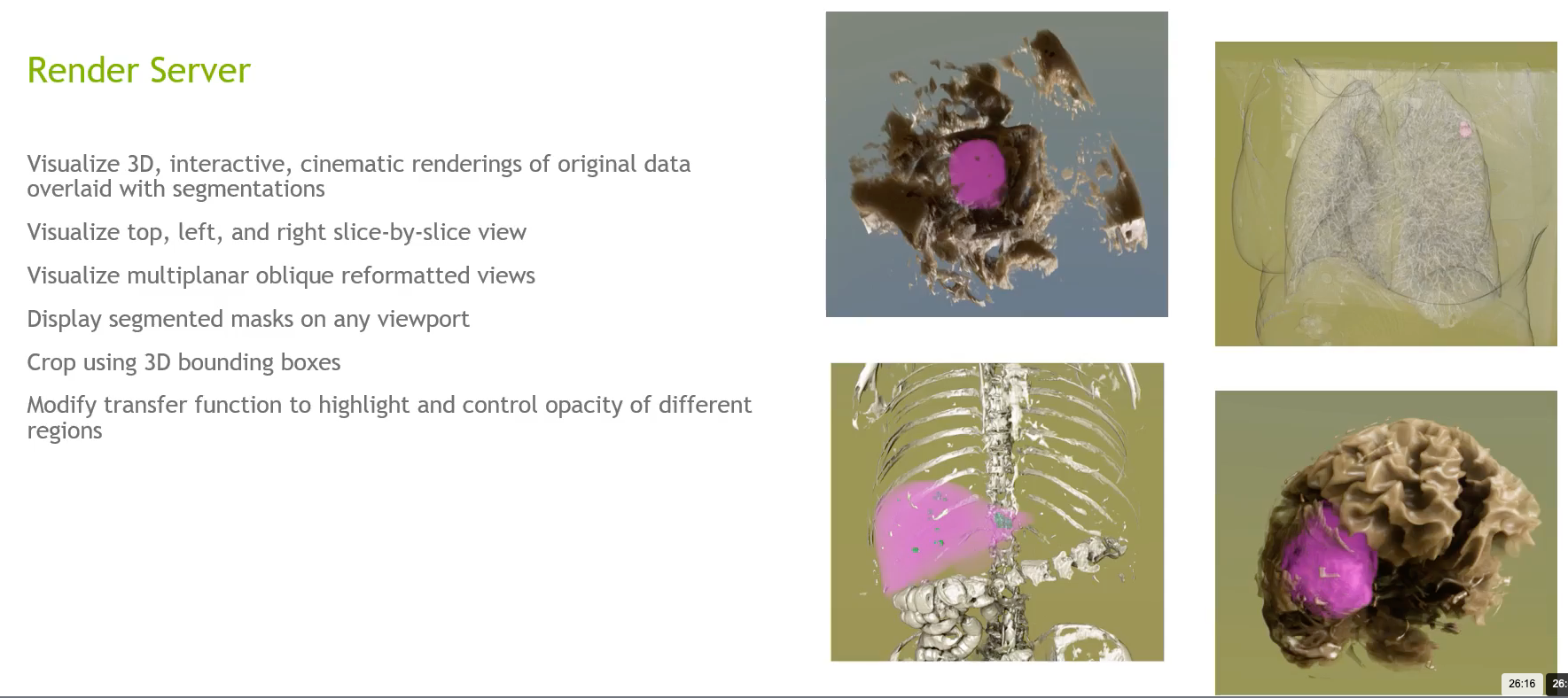
The Render Server
Nvidia Clara
Hands-on Demo
- Showcase several services of the architecture
- Use the gateway to get federated data
- Compute a model on a Nvidia Tesla Tensor Core GPU & Visualize the results
What's next?
- Federated Learning
- More models
- Add Business Logic
- Distributed Computing
- Build a scientific collaboration platform


on Github

@juliocesar_io
https://juliocesar.io
Python in life sciences, federating data for science
By Julio César
Python in life sciences, federating data for science
A data federated architecture for training AI models using Ariadne, Apollo and Nvidia Clara computing platform in Kubernetes for life sciences applications.
- 1,100



