week 06
Graph Neural Networks
Social Network Analysis
Message Passing
- Our goal is to assign our nodes meaningful coordinates (embeddings)
coordinates allow us to create decision boundaries for classification problems
- An embedding of a node should consider it's connections
i,e, nodes that share many connections should have similar embeddings
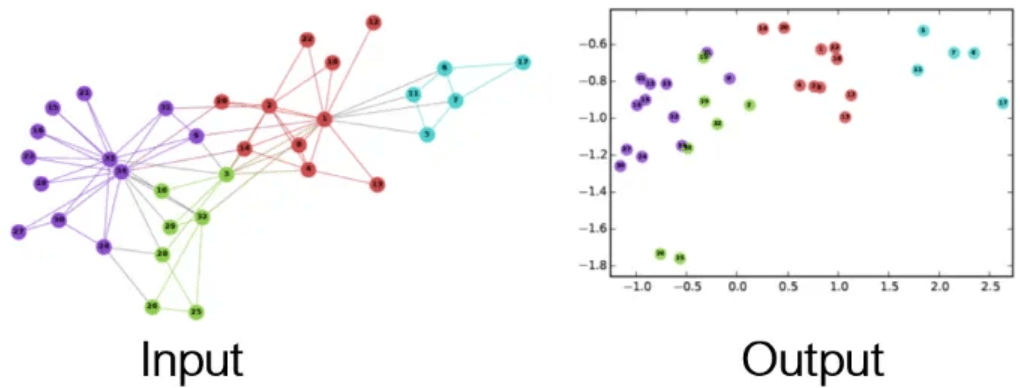
Why
Example
- Nodes: people
- Node features: age, net worth
- Edges: in phone contacts
- Edge features: number of phone calls in last year
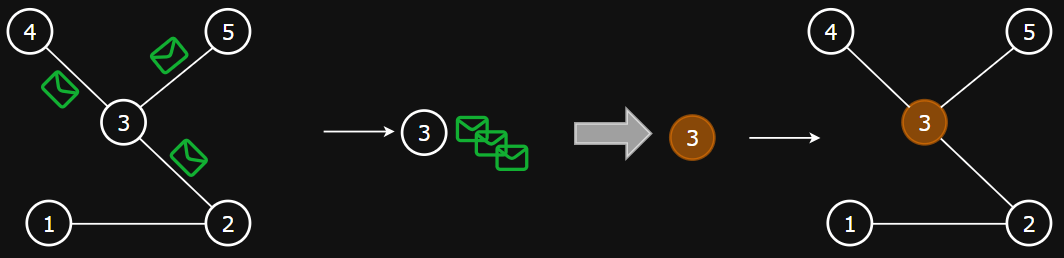
Intuition
- Goal: to calculate neighborhood-aware embeddings for nodes
- Approach:
- Messages are sent between nodes via the edges
- Nodes use these messages to update its embedding
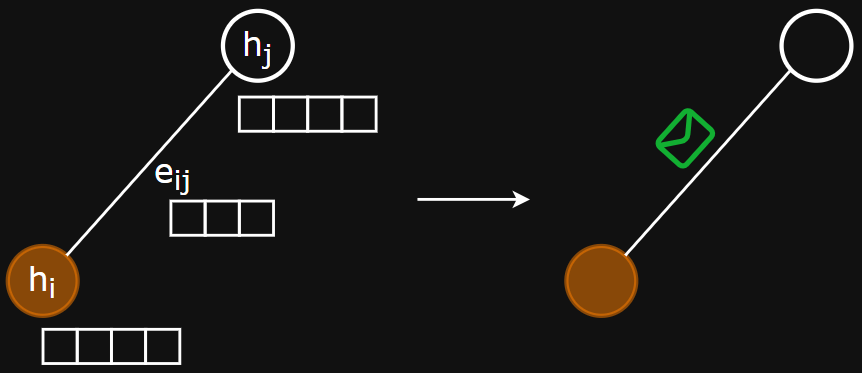
Framing the problems
- Message function - computes the message using node/edge features
- Aggregation function - combines the set of messages into a fixed-length vector that represents the neighbourhood
- Update function - computes new node embeddings using aggregated messages and the old node embedding
Message function

Message function examples
Aggregation function
aggregate all the messages from the neighborhood of i
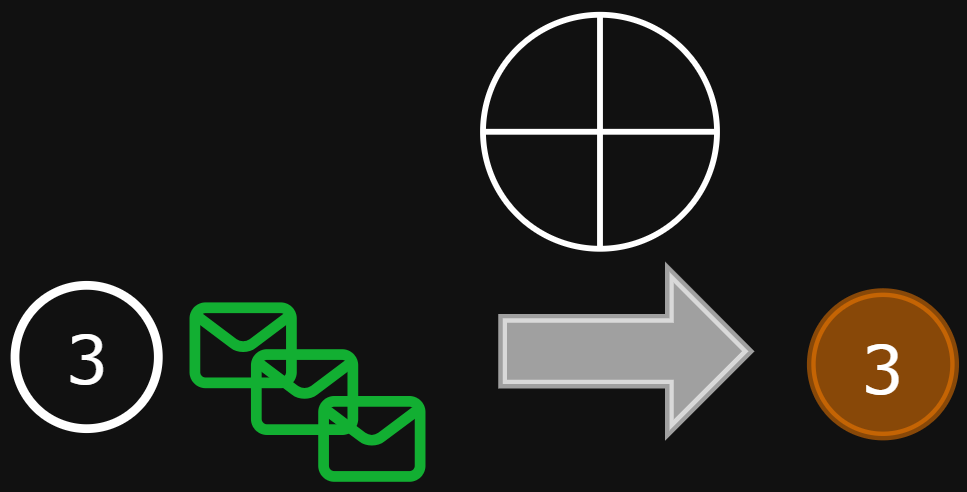
Aggregation function properties
- Fixed-lentgh representation regardless of neighborhood size
- Permutation invariant: gives the same answer regardless of how you order the inputs
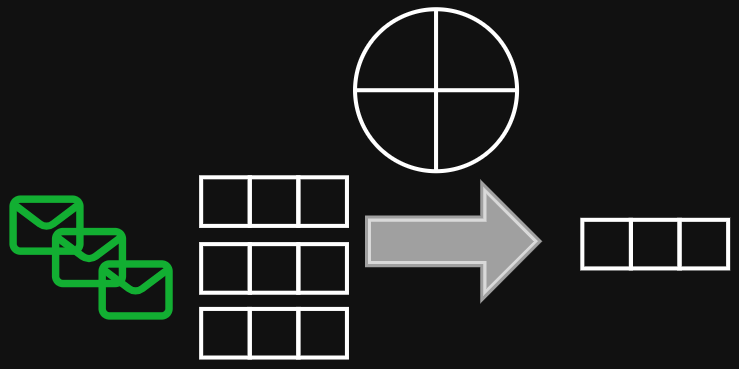

Aggregation function examples
Update function
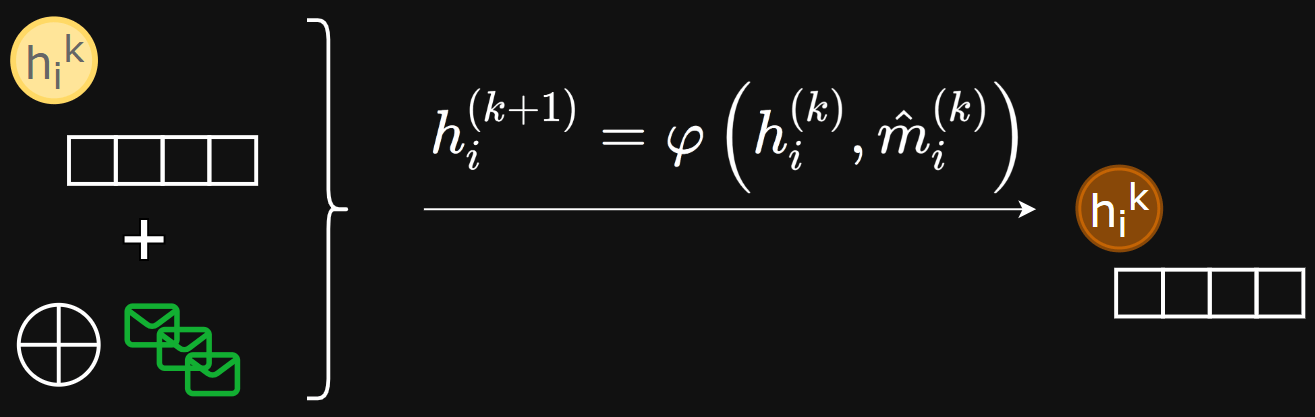
Update function examples
Architecture examples - GCN
Examples in code - GraphSage
Graph Attention Networks
Examples
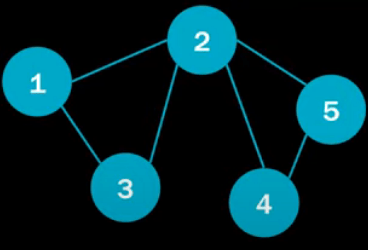

Examples

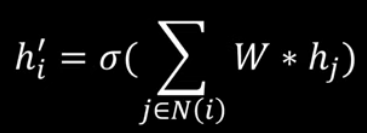
Examples


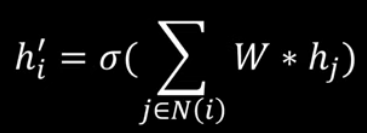


Examples


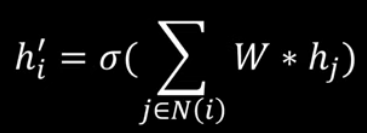


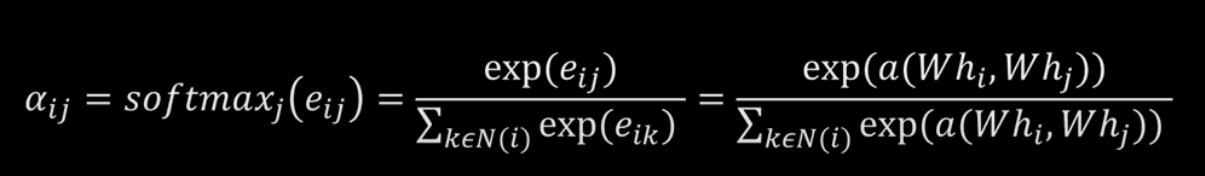
Graph Classification and Regression Overview
Examples
- Predict if this molecule:
- Is toxic
- Activates a protein / Treats a disease
- General properties of molecules
- Predict the biological taxonomy of a protein interaction network
- What type of object a point cloud represents
- The expression made by a face, represented as a mesh
Overall Architecture
- Input features X for nodes and edges, when available
- OPTIONAL: Preprocess features by passing through an MLP, shared across the nodes
- Stack GNN layers to incorporate graph neighborhood information and get node embeddings
- Use a Readout function to pool node-level embeddings into a graph-level embedding
- Use the graph-level embedding to make a prediction (e.g., with an MLP) on the graph
- Loss functions are the standards: cross-entropy for classification tasks, mean-squared error for regression
Graph Pooling
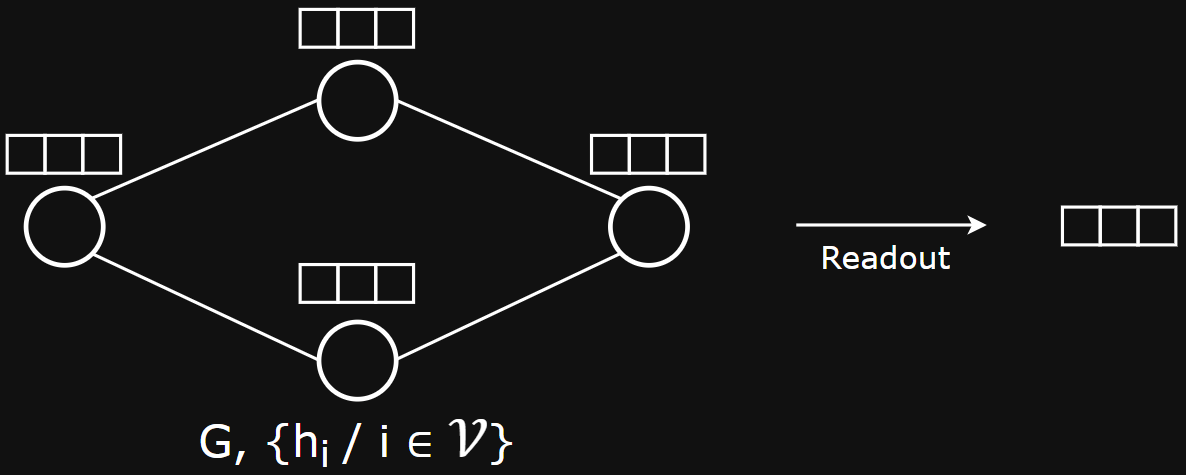
Graph Pooling with Set Pooling
- Set pooling: map a set of embeddings to a single embedding
- I.e.,
- Does not consider graph topology
- Sets do not have a natural order and the operation should therefore be Permutation Invariant
- Can use the same Permutation Invariant functions we used when creating node-neighborhood representations in GNNs: aggregation functions like SUM, MEAN, MAX, …, etc.
- E.g.,
Graph Pooling with Coarsening
- Iteratively down-samples, typically by clustering nodes and representing the cluster by a single embedding
- For each iteration, the adjacency matrix is changing
- Clustering operation needs to be differentiable so operation can be used end-to-end
- E.g. Graph U-Nets which projects nodes into 1-dimension using learnable linear layer and chooses k-largest ones as subset for next iteration.
H. Gao, S. Ji: Graph U-Nets [link]
Nuances of Graph Batching
- In previous tasks, we had one large graph and were sampling nodes and their k-hop neighborhoods
- Now: 1 sample = 1 full graph, no edges between graphs in the batch
- How should we do message passing?
- Could loop over each graph and run MP separately (slow)
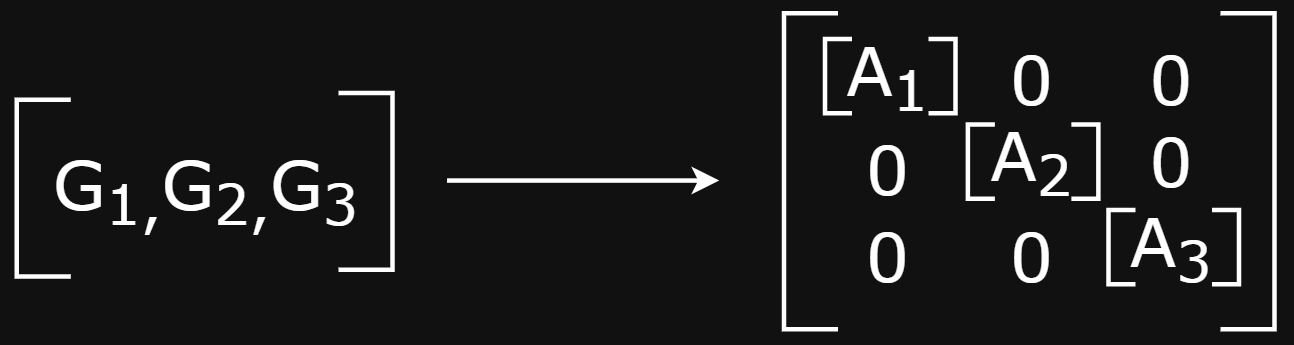
The batched “super-graph”
- Alternatively, could create a “super-graph” that creates a block-diagonal adjacency matrix of disconnected components and run MP once (fast)
- This creates book-keeping complexity of keeping track of which node belongs to which graph. This is important when need to do graph pooling to get graph-level embeddings
- This is DGL’s approach and they provide tooling for managing this complexity (e.g., Batching and Reading Out Ops)
Batching and Reading Out Ops [link]
The batched “super-graph”
Graph Classification Exercise
- ogbg-molhiv [link] dataset from Open Graph Benchmark:
- Predict whether a molecule (graph) inhibits HIV replication
- 41k molecules from MoleculeNet [link]
- Average of 26 nodes and 28 edges per graph
- Natural features for both atoms (nodes) and bonds (edges):
- Categorical in nature and the features are therefore integers that indicate the category mapping for each column
- List of features is here, but include things like atomic number, bond type, …, etc.
- OGB provides classes for converting these categorical/integer features into embeddings: AtomEncoder [link] and BondEncoder [link]
- Split into train/val/test based on molecular structure
- To evaluate generalization, the test nodes have structural differences from those in the training set
Practice in DGL
- Started as research project out of NYU Shanghai
- Joined forces with AWS and is now developed/maintained there in collaboration with NVIDIA and the open source community
- Differentiated by speed and scalability
- Supports multi-GPU and multi-machine with multi-GPU
- Documentation has nice section of tutorials for learning and there are many examples in github
- Backend is framework agnostic
Deep Graph Library (DGL)
DGL 1.0 release
dgl blog [link]
DGL 2.0 release
dgl blog [link]
Useful APIs and data structures
- Named node and edge features
- g.ndata['x']=X
- g.edata['a']=E
- Graph processing
- dgl.add_reverse_edges(g)
- dgl.add_self_loop(g)
- Graph querying
- g.num_nodes()
- g.num_edges()
- g.has_edges_between(u,v)
- g.in_degrees()
- \( h_u=\frac{1}{N_u}\sum_{V \in N_u}x_v \) = g.update_all(fn.copy_u('x', 'm'), fn.mean('m','h'))
- Message passing APIs
- Increasingly supporting heterogeneous graphs
PyTorch Geometric (PyG)
Built on top of PyTorch
- Developed at TU Dortmund and Stanford Universities
- Overlap with group that runs the Open Graph Benchmark
- Standardizes the API around defining “message” and “update” functions, along with specifying an aggregator
- Has a large set of built-in datasets and implementations
New(er) comers
- Jraph
- Comes from DeepMind
- Built on top of JAX
- TensorflowGNNs
- Alpha release announced Nov 2021
- Keras-style API
- A stated emphasis on heterogeneous graphs
Open Graph Benchmark
- Larger and more realistic benchmark graph datasets
- Node, Link and Graph property prediction tasks
- Wrapped in a Python package for easy loading into PyGand DGL
- Comes with a pre-defined train/val/test split
- Also comes with built in “Evaluators” for each dataset to ensure performance is consistently measured
- Each dataset has a leaderboard, each row is a method’s performance with paper + code
- There’s also a “Large-Scale” version with much larger graphs
OGB [link]
- Wang, M., Zheng, D., Ye, Z., Gan, Q., Li, M., Song, X., ... & Zhang, Z. (2019). Deep graph library: A graph-centric, highly-performant package for graph neural networks [pdf]
- Fey, M., & Lenssen, J. E. (2019). Fast Graph Representation Learning with PyTorch Geometric Computer software [link]
- Godwin*, J., Keck*, T., Battaglia, P., Bapst, V., Kipf, T., Li, Y., Sanchez-Gonzalez, A. (2020). Jraph: A library for graph neural networks in jax(Version 0.0.1.dev). Opgehaalvan [link]
- Hu, W., Fey, M., Zitnik, M., Dong, Y., Ren, H., Liu, B., ... & Leskovec, J. (2020). Open graph benchmark: Datasets for machine learning on graphs [pdf]
Extra - Improving Scalability
The problem(s)
- Many real-world/industrial graphs are billions of nodes and edges, perhaps 100s of billions
- E.g., E-commerce products, reviews, transactions, sign-in events
- You may not even be able to fit this into memory
- In small-world graphs like social networks, ~6 GNN layers means every node in the graph is needed to calculate each node embedding
Sampling
- Idea: maybe you don’t need the full graph to compute useful embeddings
- In fact, having this source of randomness may make your model generalize better to new, slightly different neighborhoods
- Can sample at different levels:
- Subset of neighbors of each node during message passing
- A set of nodes for each GNN layer
- A subset of the graph for the full GNN
Node-level Sampling
- Random neighbor sampling: for each node, only choose a subset of neighbors for message passing (e.g., GraphSAGE)
- Still suffers from exponential growth with layers, but controllable
- Importance sampling: try to improve the variance of your estimate by smarter sampling
- FastGCN and LADIES use importance sampling at the layer-level to reduce exponential growth to linear growth.
W. L. Hamilton, R. Ying, J. Leskovec: Inductive Representation Learning on Large Graphs [link]
J. Chen, T. Ma, C. Xiao: FastGCN: Fast Learning with Graph Convolutional Networks via Importance Sampling [link]
D. Zou, Z. Hu, Y. Wang, S. Jiang, Y. Sun, Q. Gu: Layer-Dependent Importance Sampling for Training Deep and Large Graph Convolutional Networks [link]
Subgraph sampling
- Cluster-GCN: Find densely connected clusters and only sample neighbors in the same cluster
- GraphSAINT: Create random subgraphs for each minibatch and model with a GCN as if it were the full graph
Pre-computation
- Idea: instead of doing message passing before each MLP layer, do graph-based processing once
- Assumes that the non-linearity between GNN layers is not important, so the model fitting phase is ~ logistic regression
- Can be done in one big batch-processing job, not during training process
- Simple Graph Convolution (SGC): \( H = softmax(A^KX\theta) \)
- Scalable Inception Graph Neural Networks (SIGN): Similar to SGC, but also consider general operators, not just powers of A (e.g., counting triangles, diffusion operators like Personalized Page Rank,..., etc) and concatenate results as features
M. Chen, Z. Wei, Z. Huang, B. Ding, Y. Li: Simple and Deep Graph Convolutional Networks [link]
F. Frasca, E. Rossi, D. Eynard, B. Chamberlain, M. Bronstein, F. Monti: SIGN: Scalable Inception Graph Neural Networks [link]
H. Wang, Z. Wei, J. Gan, S. Wang, Z. Huang: Personalized PageRank to a Target Node, Revisited [pdf]
Resource management
- There are optimizations that can be made to improve resource utilization
- GNNAutoScale caches historical embeddings from previous minibatches to avoid re-computing them
- ROC and DistGNN enable fast/scalable distributed training via memory and graph partitioning optimizations
M. Fey, Jan E. Lenssen, F. Weichert, J. Leskovec: GNNAutoScale: Scalable and Expressive Graph Neural Networks via Historical Embeddings [link]
Z. Jia, S. Lin, M. Gao, M. Zaharia, A. Aiken: Improving the accuracy? scalability, and perfomance of graph neural networks with ROC [pdf]
V. Md, S. Misra, G. Ma, R. Mohanty, E. Georganas, A. Heinecke, D. Kalamkar, N. K. Ahmed, S. Avancha: DistGNN: Scalable Distributed Training for Large-Scale Graph Neural Networks [link]
Deep Graph Library (DGL)
By karpovilia
Deep Graph Library (DGL)
- 293



