Domain inspired machine learning for hypothesis extraction in biological data
Karl Kumbier
UC Berkeley Statistics
Advisor: Prof. Bin Yu
In search of mechanistic insights into development and function

- Genomes contain vast amounts of information (e.g. 6M variants in 20K human genes)
- Complex biological processes are the result of interacting components (e.g. 1e170 potential interactions among human variants)
- Linking genomic processes to biological outcomes requires the exploration of enormous spaces
The PCS framework: evaluating human judgement calls in data science
- Predictability: Does my model reflect external reality?
- Computability: Can I tractably build/train my model?
- Stability: Are my results consistent with respect to "reasonable" perturbations of the data/model?
Outline
-
From genomic to statistical interactions
-
Market baskets and genomics
-
Iterative Random Forests
-
Interaction discovery in Drosophila
From genomic to statistical interactions
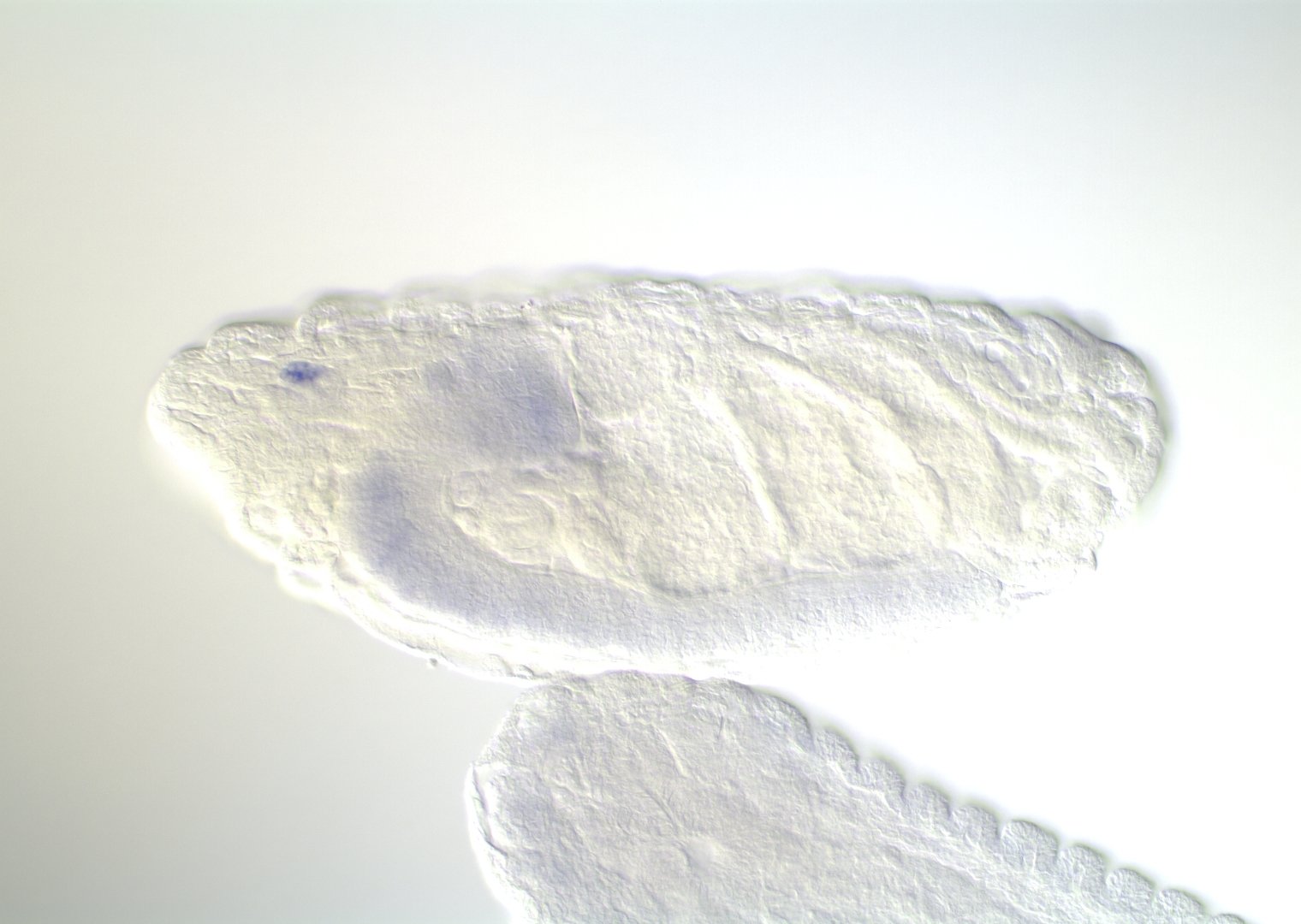
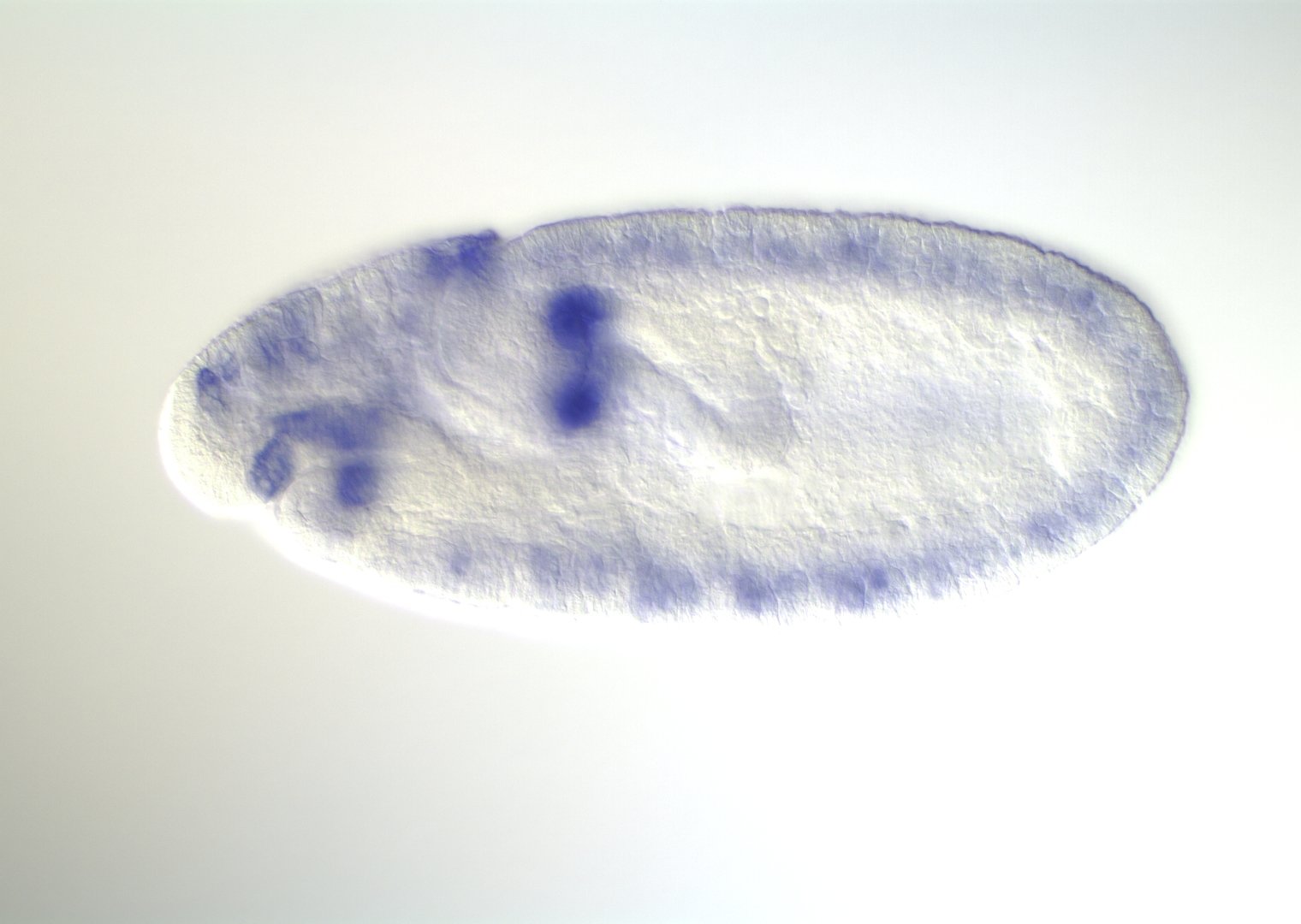
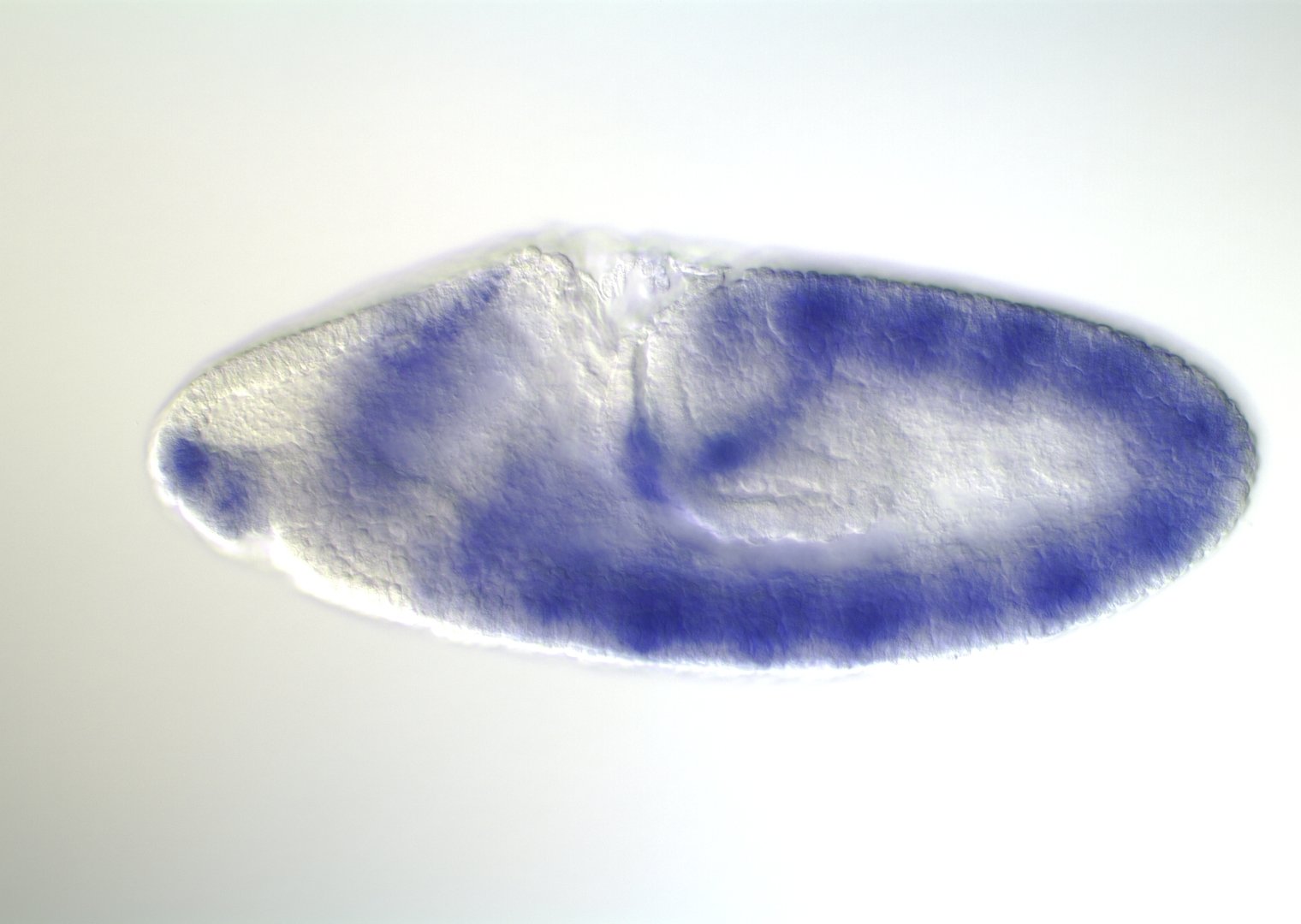
Embryonic development in Drosophila
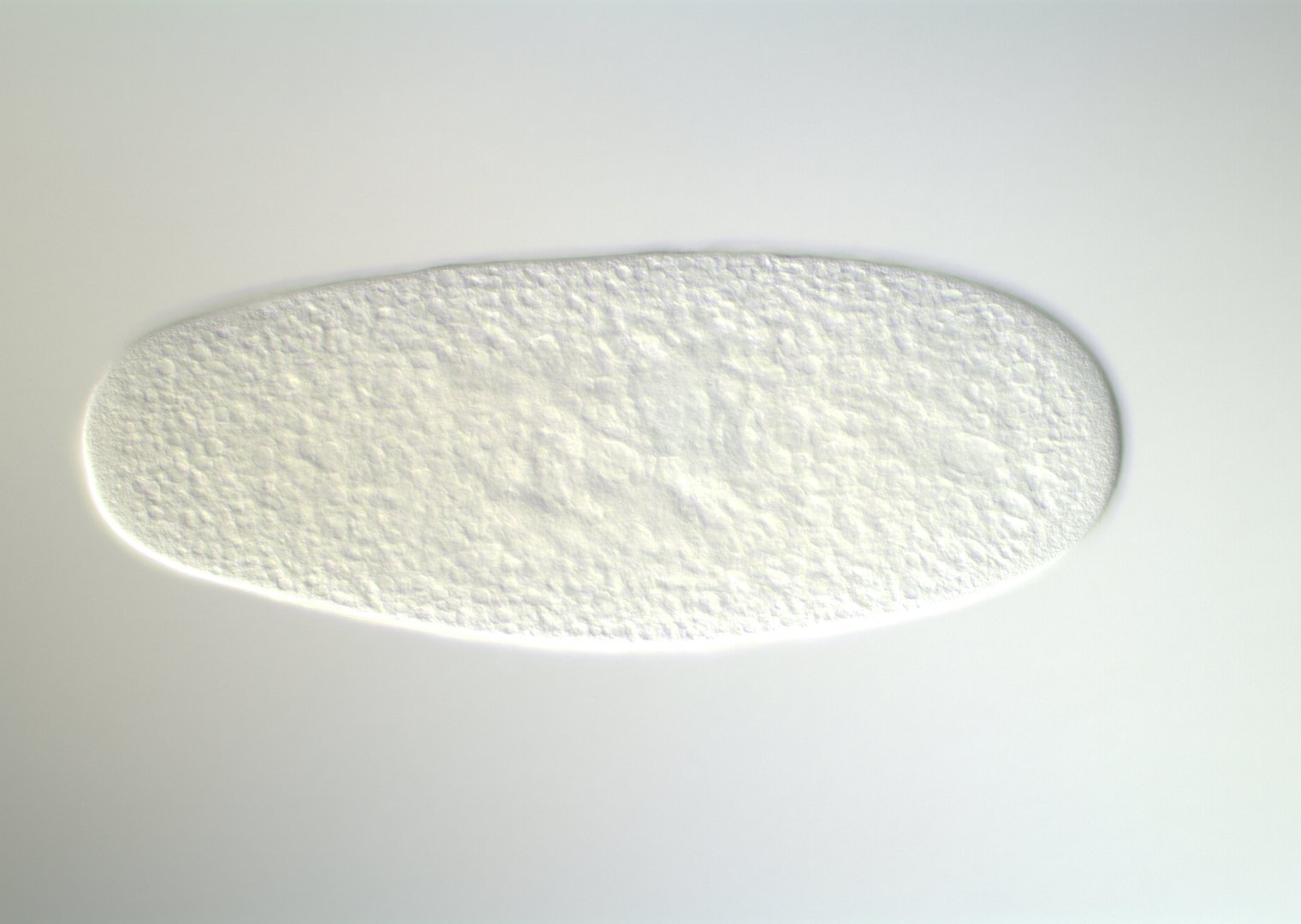
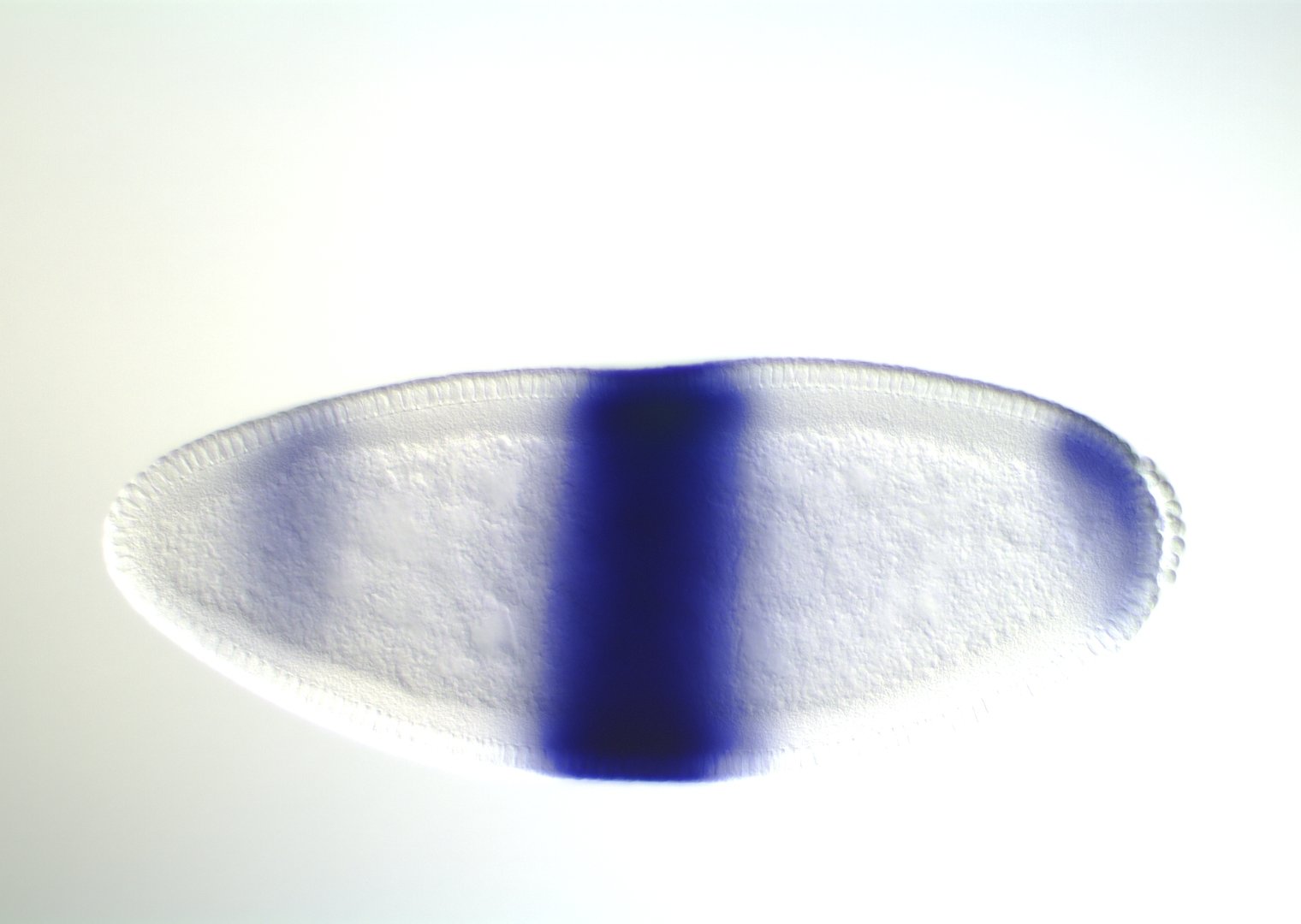
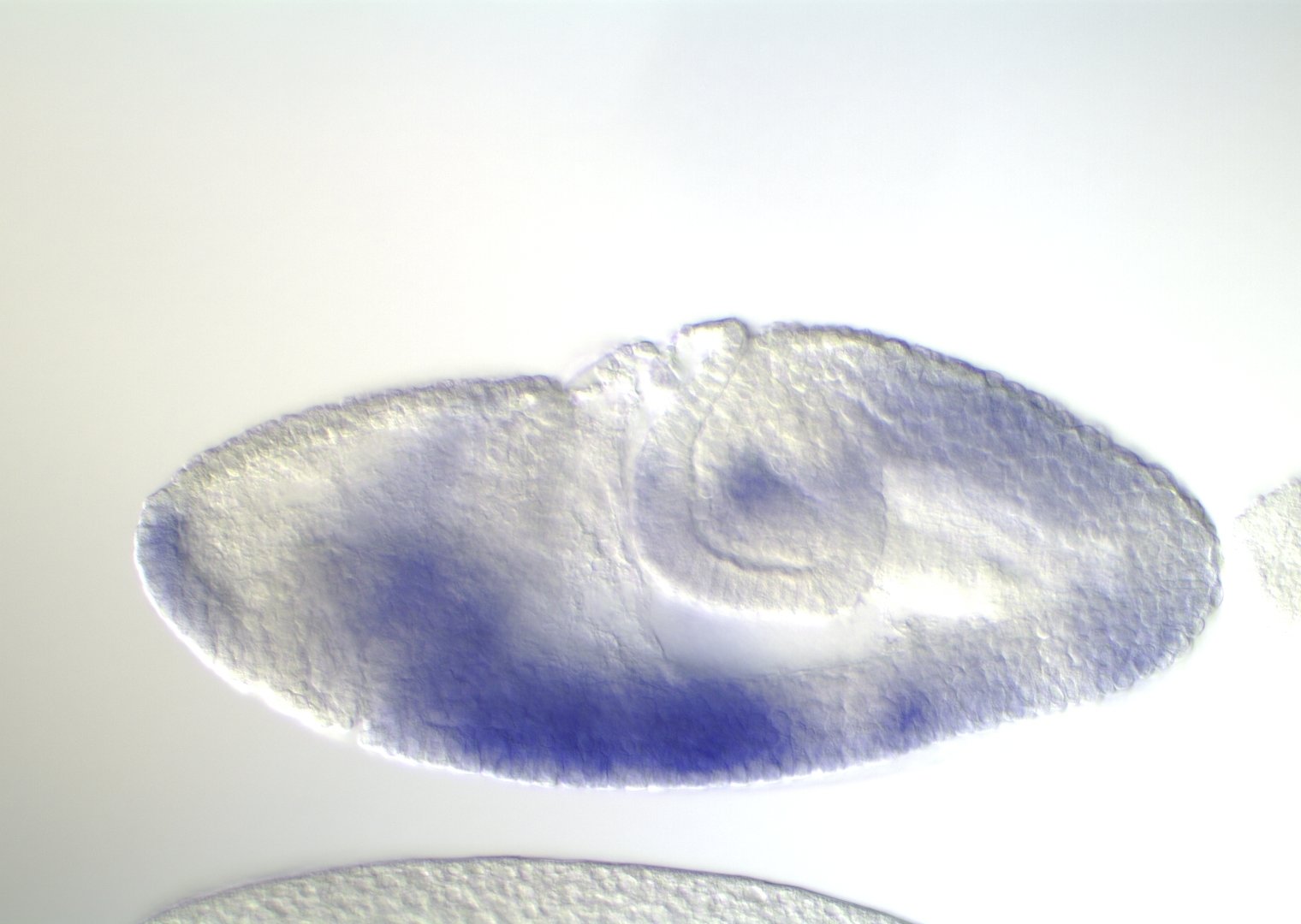
0-1:20 hours
1:20-3:00 hours
3:00-3:40 hours
3:40-5:20 hours
5:20-9:00 hours
9:20-16:00 hours
image: Volker Hartenstein
images: BDGP
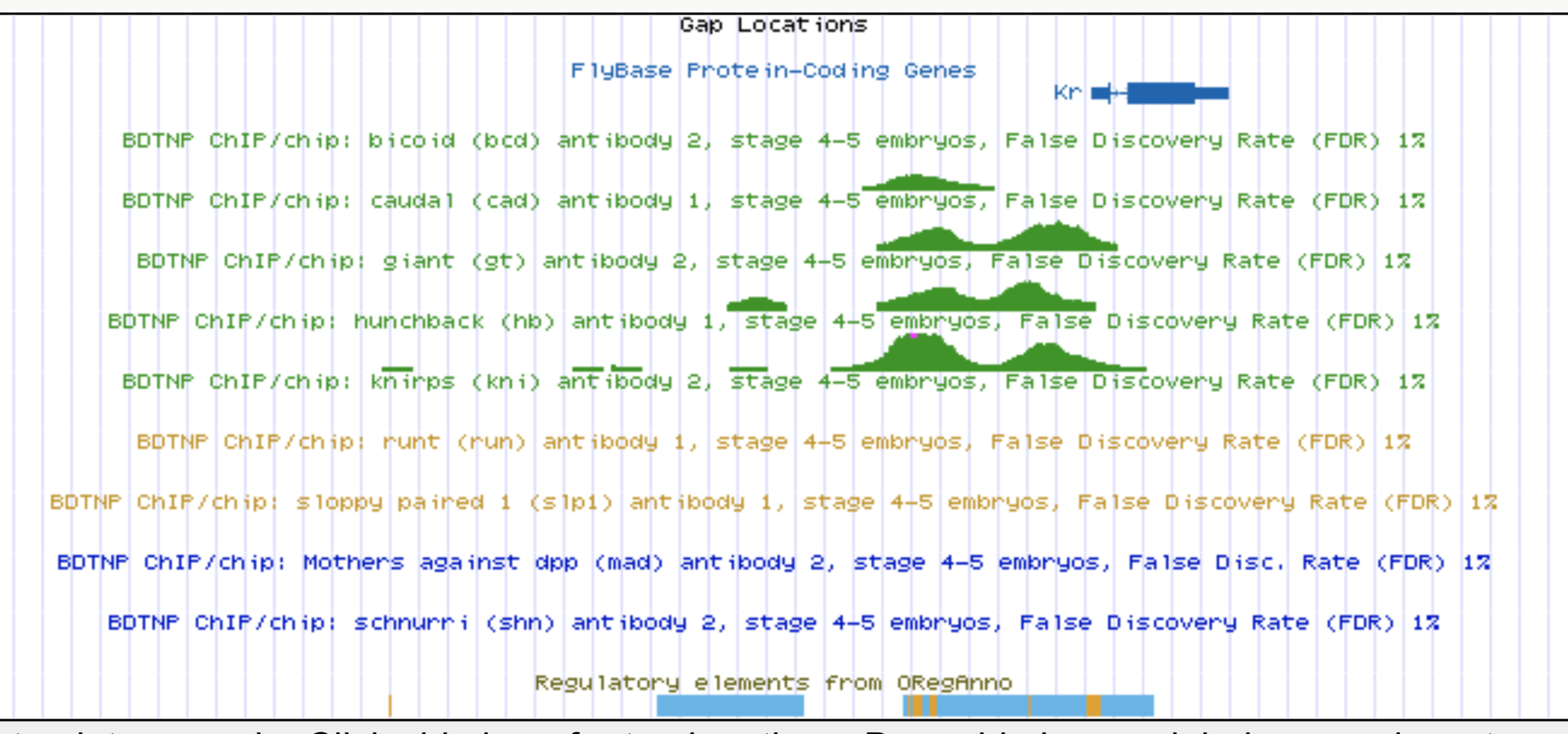
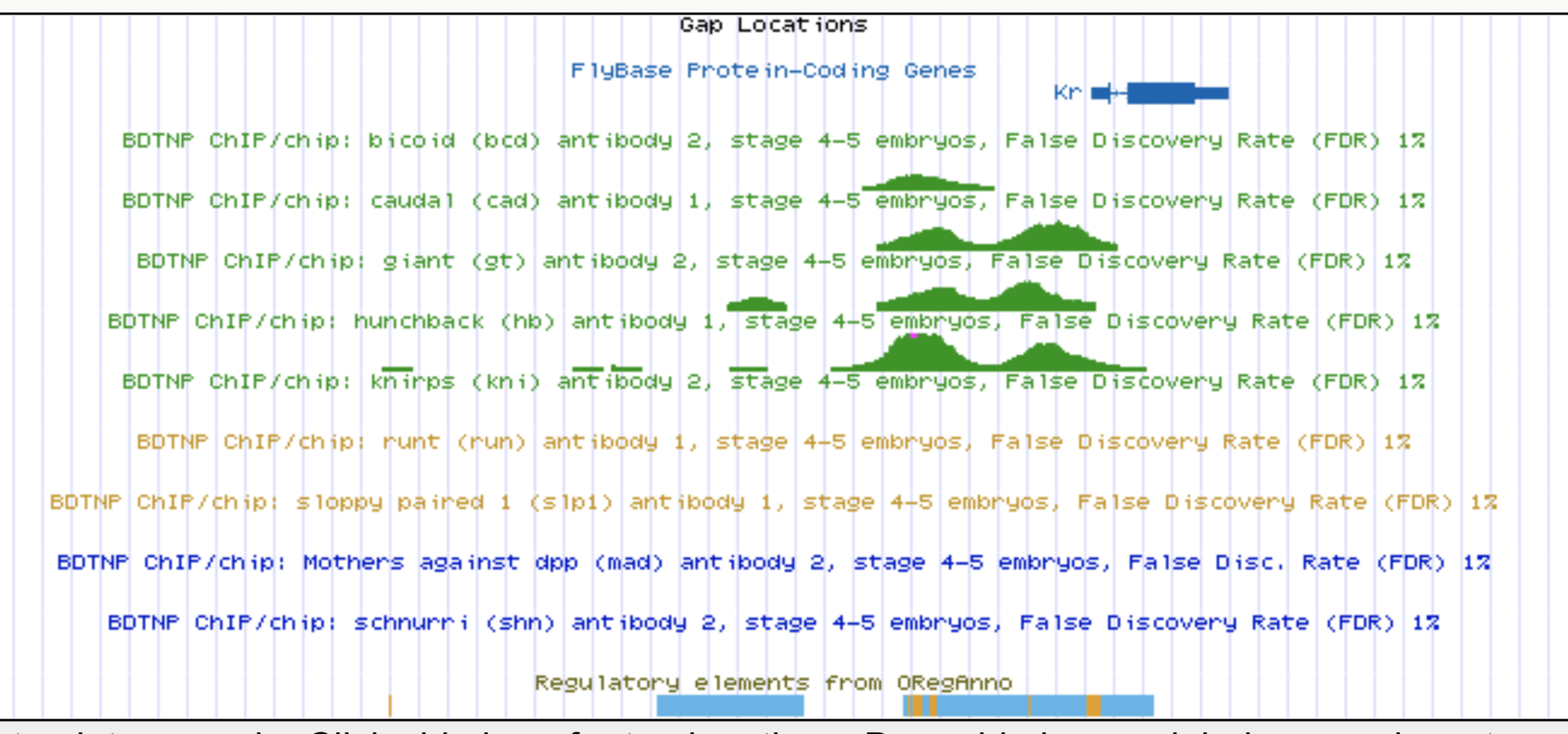
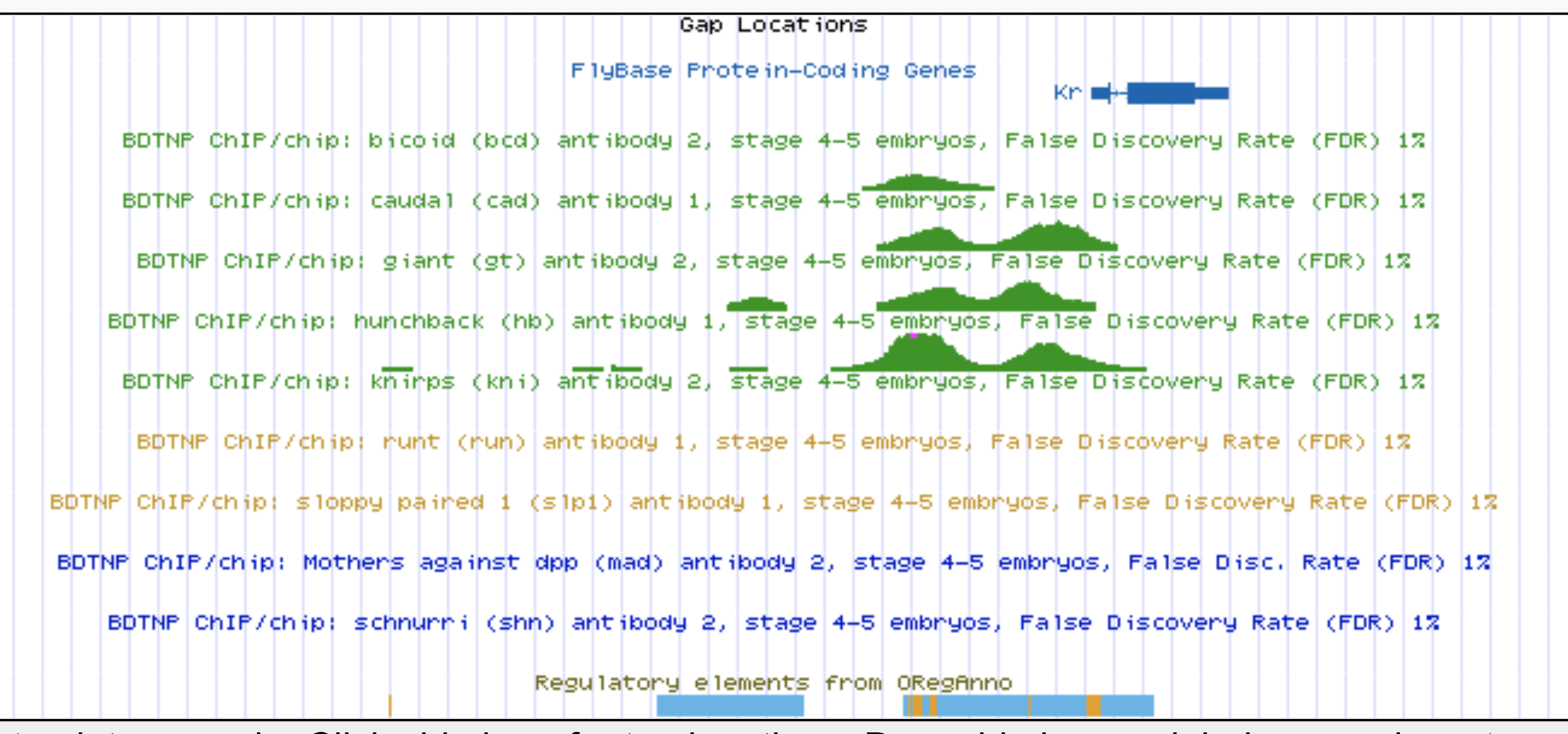
Kr expression
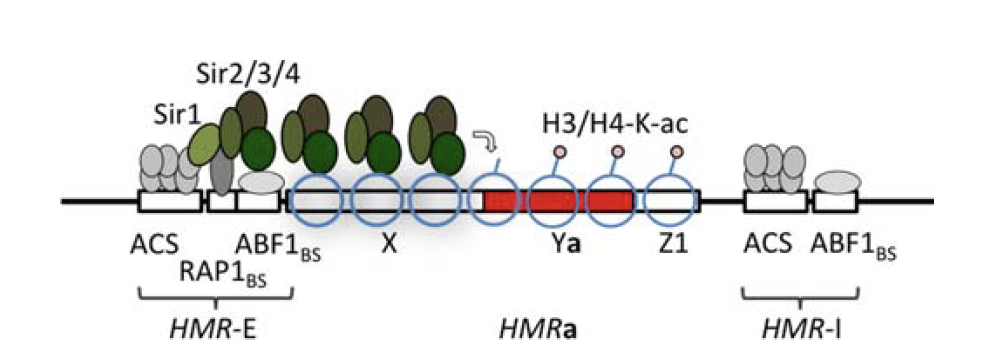
Enhancers regulate spatio-temporal programs of gene expression
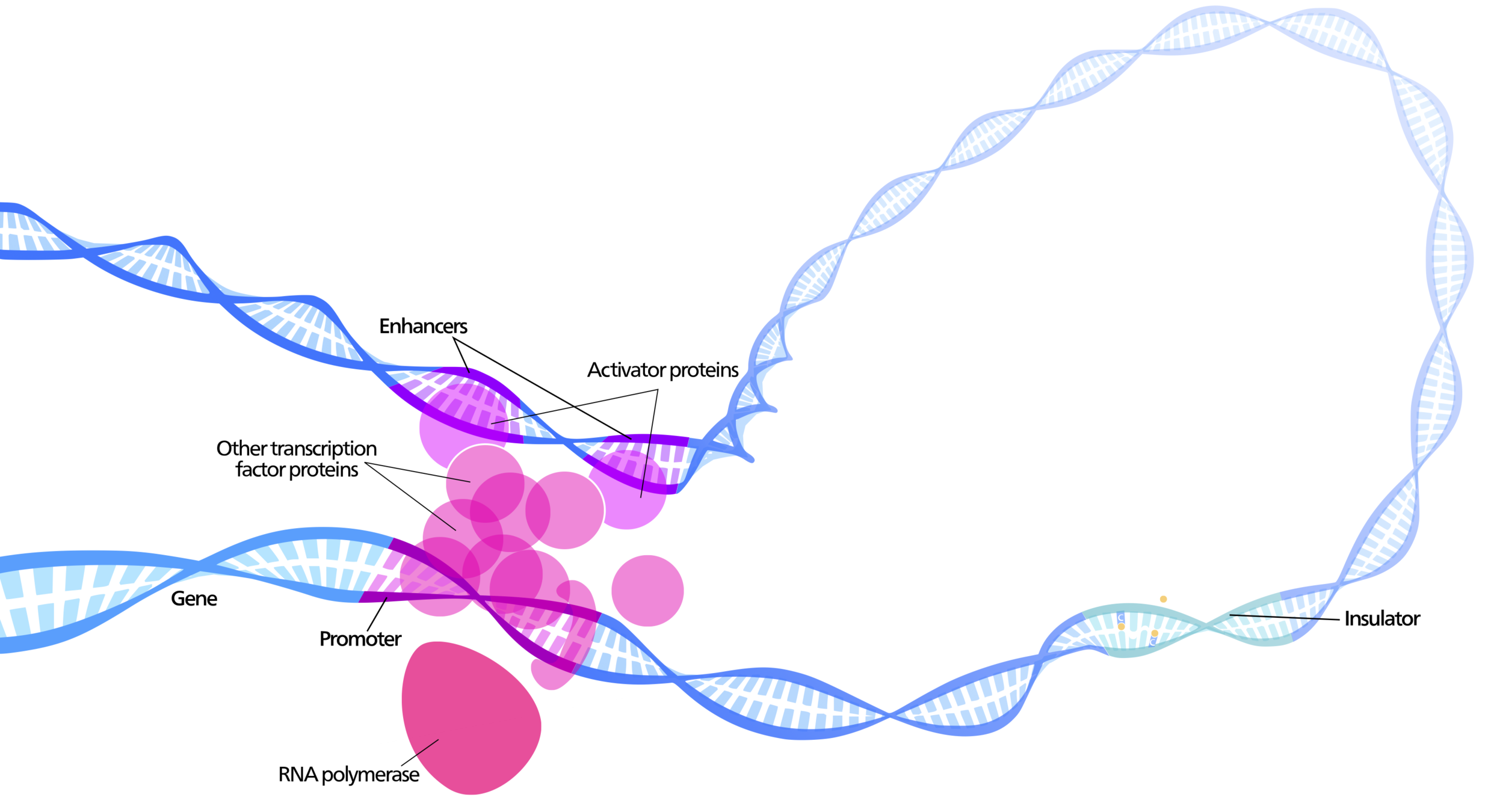
Enhancers: segments of the genome that coordinate transcription factor (TF) activity to regulate gene expression.
Experimental evaluation of enhancer elements in Drosophila
Pfeiffer et al. (2008)
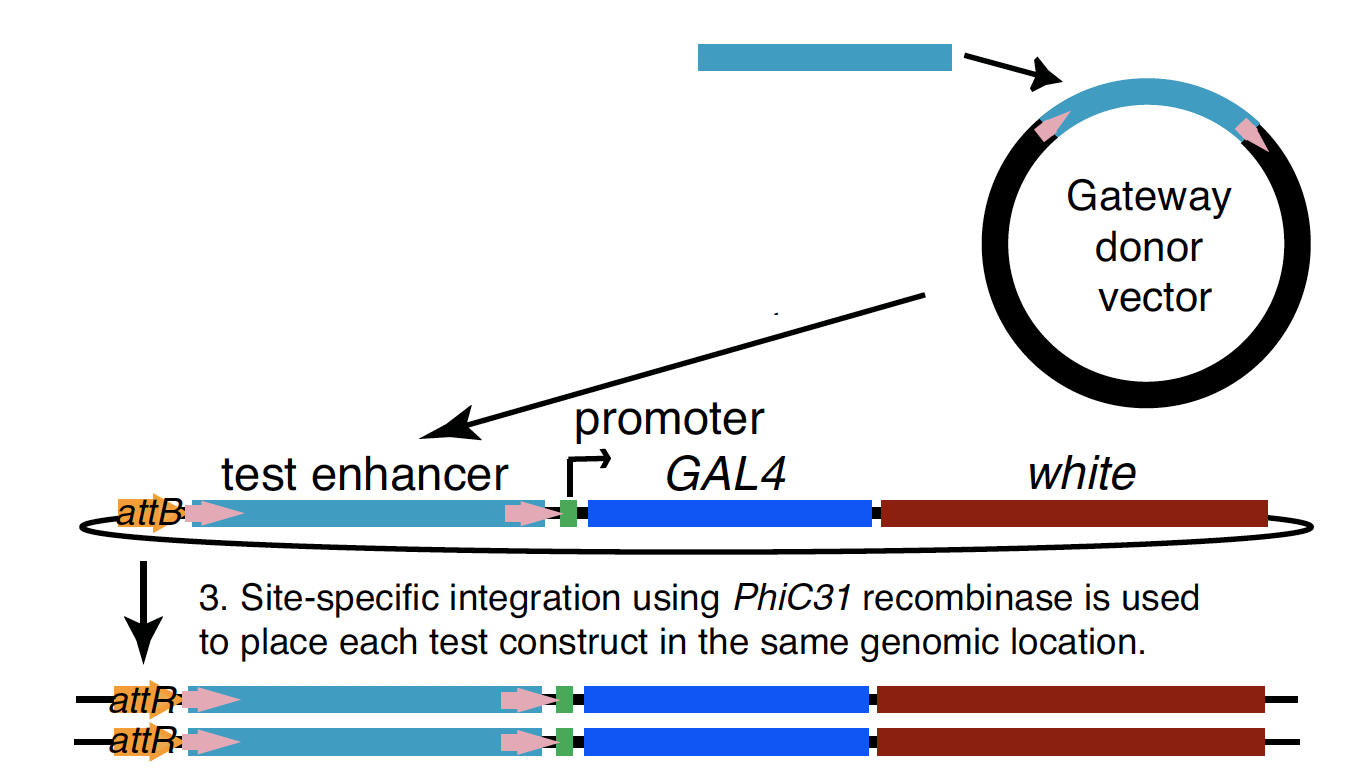
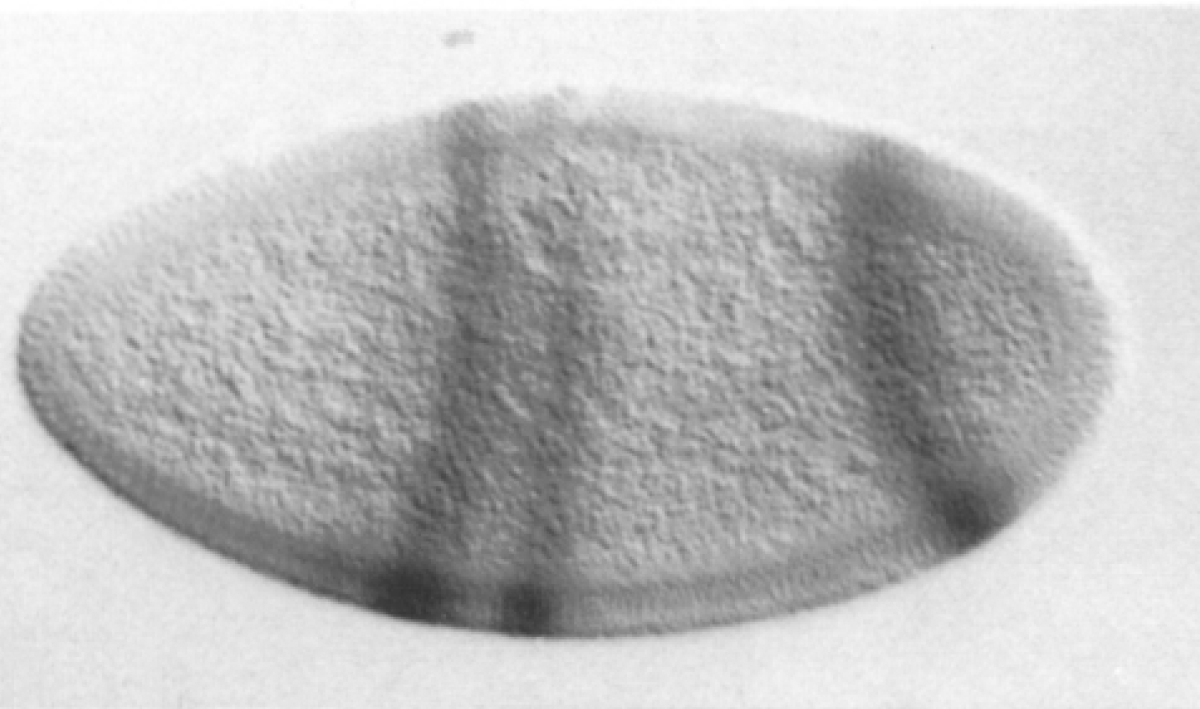
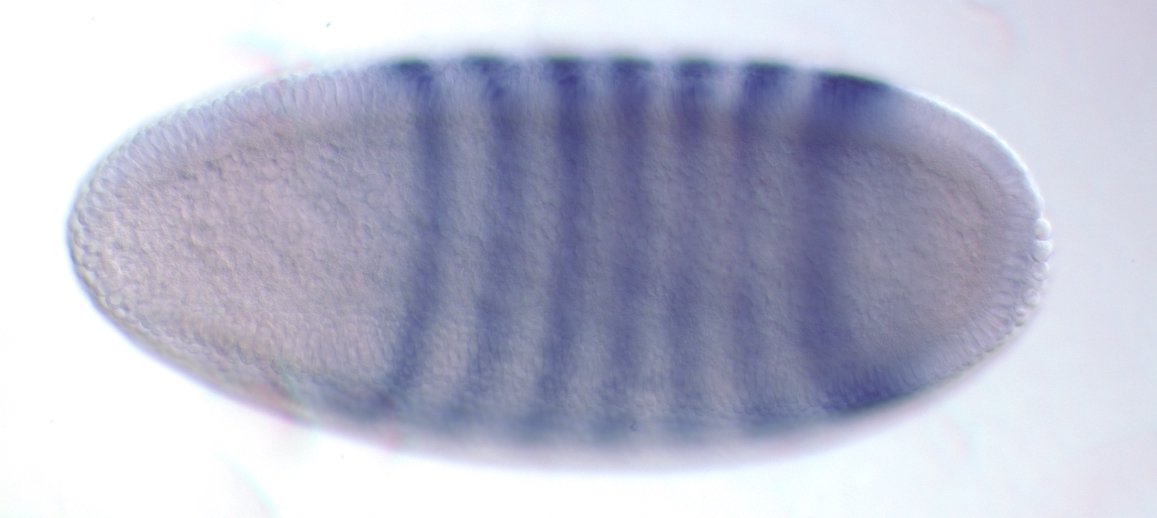
even-skipped
expression
wt
transgenic
Experimental evaluation of enhancer elements in Drosophila
Hiromi et al. (1985), Harding et al. (1989), Goto et al. (1989), Pfeiffer et al. (2008)



even-skipped expression
wt
transgenic
High-order interactions at enhancer elements drive embryonic development
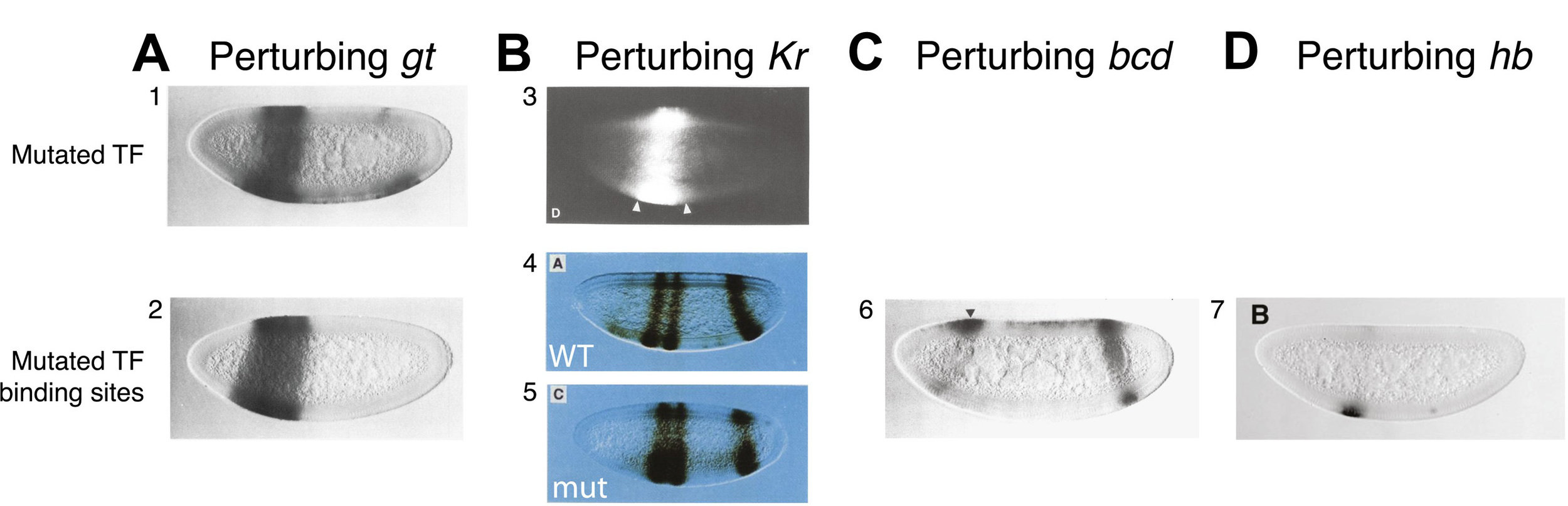
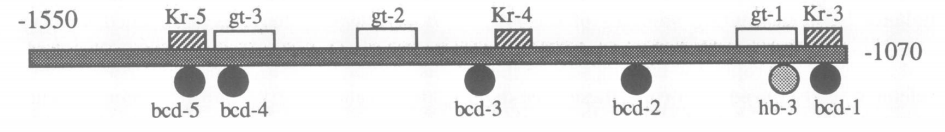
Goto et al. (1989), Harding et al. (1989), Small et al. (1992), Isley et al. (2013), Levine et al. (2013)
Identifying regulatory interactions from high-throughput genomic data
Experimentally validated enhancer elements.
Whole-embryo ChIP-chip/ChIP-seq measurements of transcription factor (TF) DNA binding
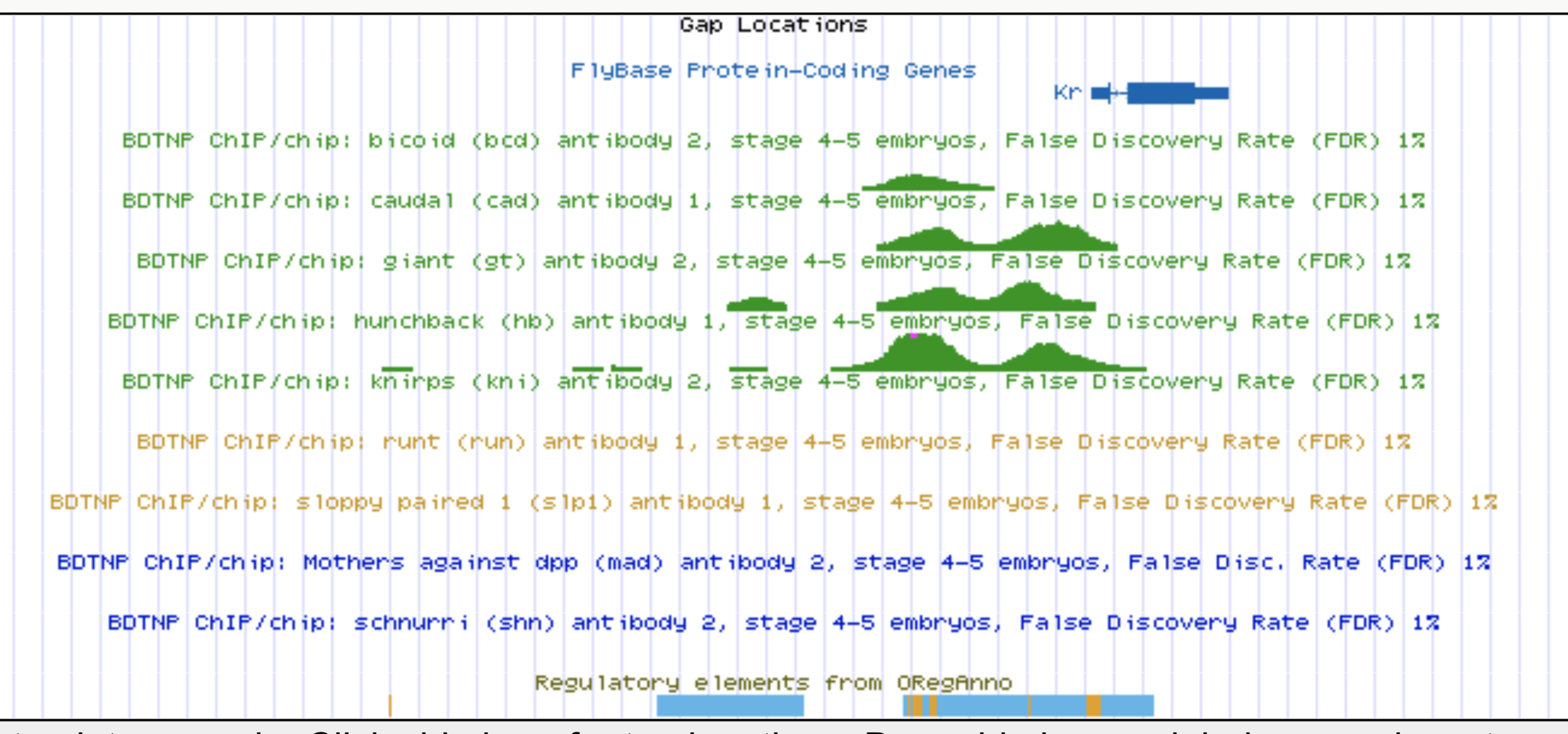
From genomic to statistical interactions
activators
repressors
Segment of the genome
DNA binding for p transcription factors (TFs)
Order-s interaction: s = #activators + #repressors
RuleFit: rule-based interaction discovery (Friedman and Popescu, 2008)
- Identify a collection of marginally important features
- Search for predictive order-2 rules among marginally important features
Computational costs grow as
Misses interactions with weak marginal effects
image: Lee and Haber (2014)
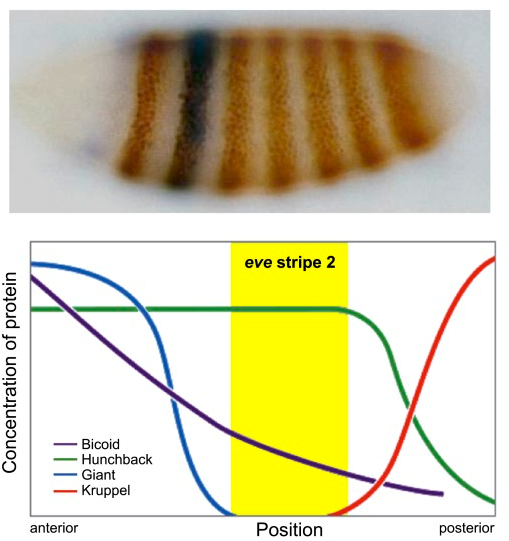
Thresholding rules define expression domains
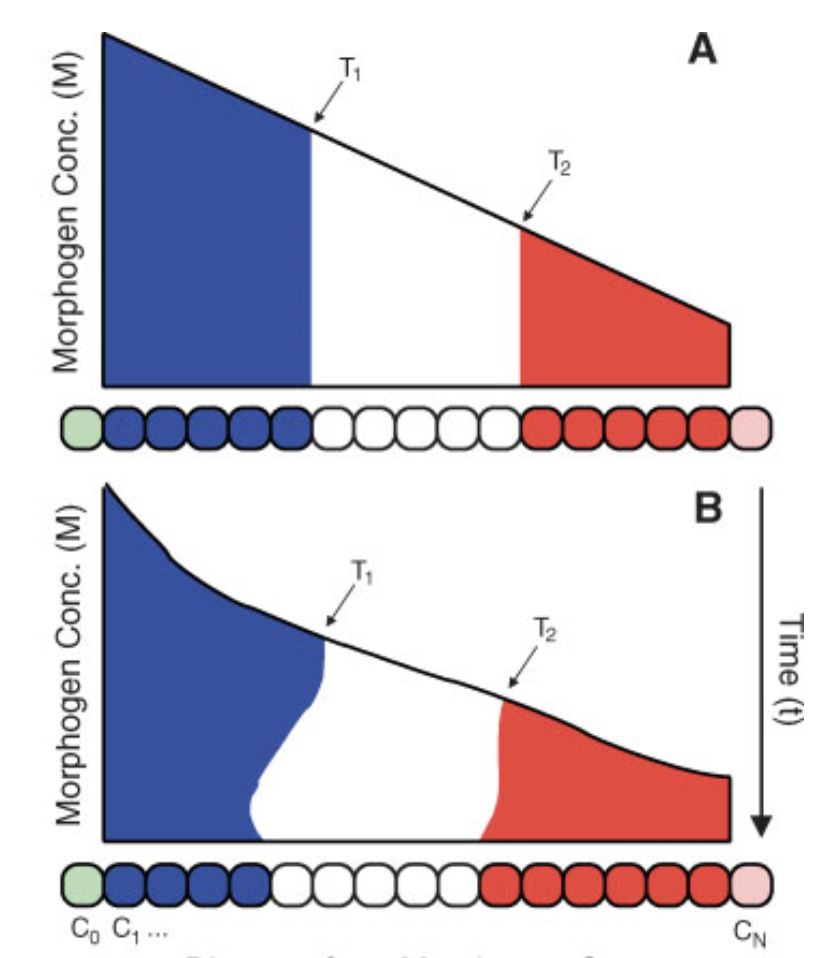
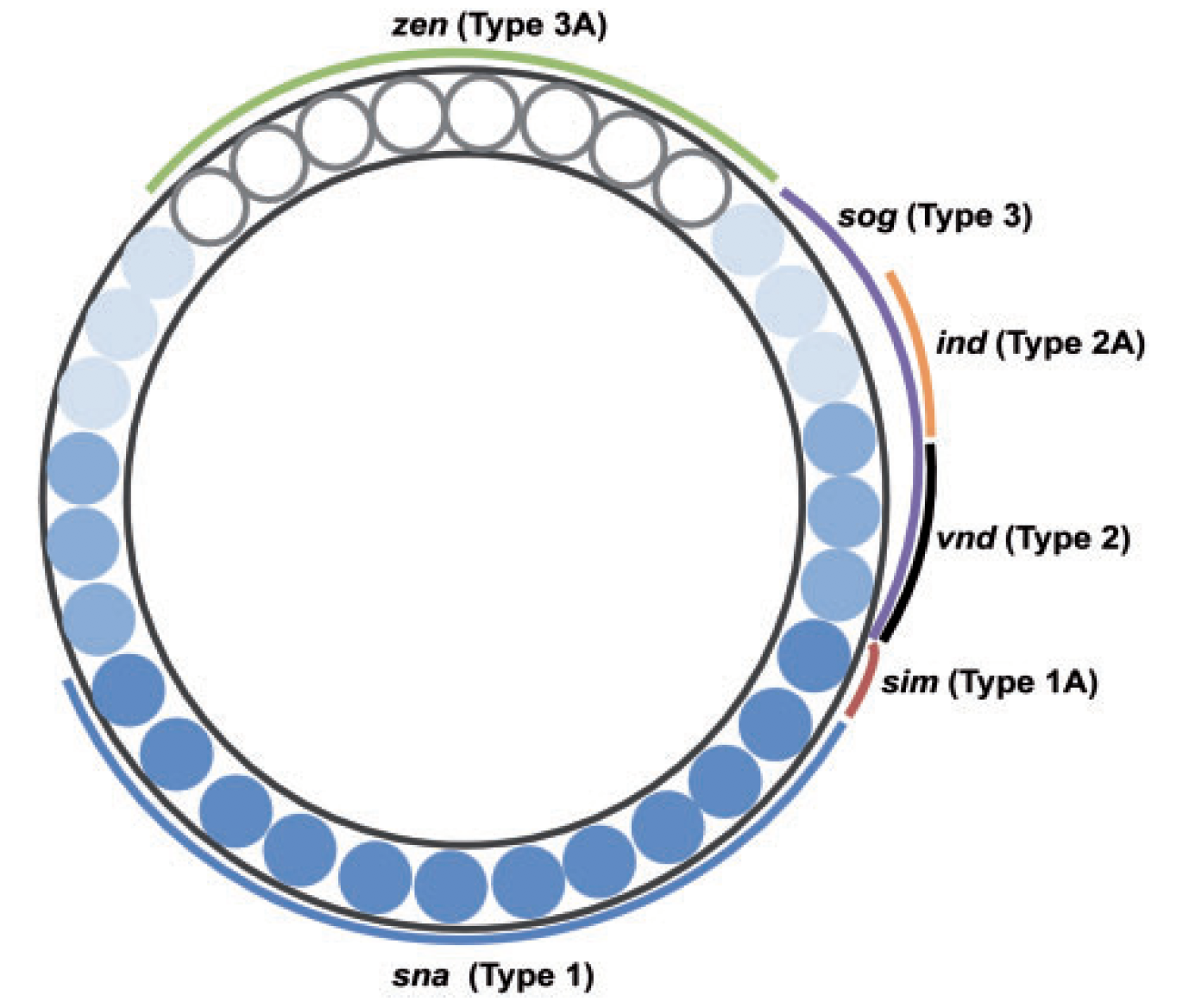
Chopra and Levine (2009)
Dl +
Dl -
Wolpert (1968), Jaeger and Reinitz (2006), Chopra and Levine (2009), Zizen et al. (2009), Knowles and Biggin (2013), Levine (2013), Staller al. (2015), ...
Jaeger and Reinitz (2009)
From genomic to statistical interactions
(1) How precisely does an interaction predict class-1 observations?
(2) How prevalent is an interaction among class-1 observations?
Interactions:
Responses:
?
Market baskets and genomics
Interactions in market baskets
















What combinations of items do customers purchase together?
Interactions in market baskets















What combinations of items do customers purchase together?
What combinations of items do different types of customers purchase together?




Interactions in market baskets



















Feature-index sets
Random intersection trees (RIT)
Shah and Meinshausen (2014)
Leverage sparsity in market baskets to search for frequently co-occurring items in a computationally efficient manner
- Randomly sample feature index sets from class-C observations:
- Intersect sampled feature index sets in a tree like fashion up to depth D
- Return all feature combinations that "survive" intersection procedure up to depth D
Random intersection trees (RIT)
Shah and Meinshausen (2014)









Randomly sampled
class-C observation
"survived" interaction
Random intersection trees (RIT)
Shah and Meinshausen (2014)




















Random intersection trees (RIT)
Shah and Meinshausen (2014)





































Genomic response
Genomic features
Translating the market basket problem into genomics



Genomic response
Genomic features
Translating the market basket problem into genomics
Challenges:
- Genomic features are typically measured in concentrations/counts
- Binding does not imply regulation (Li et al. 2008)




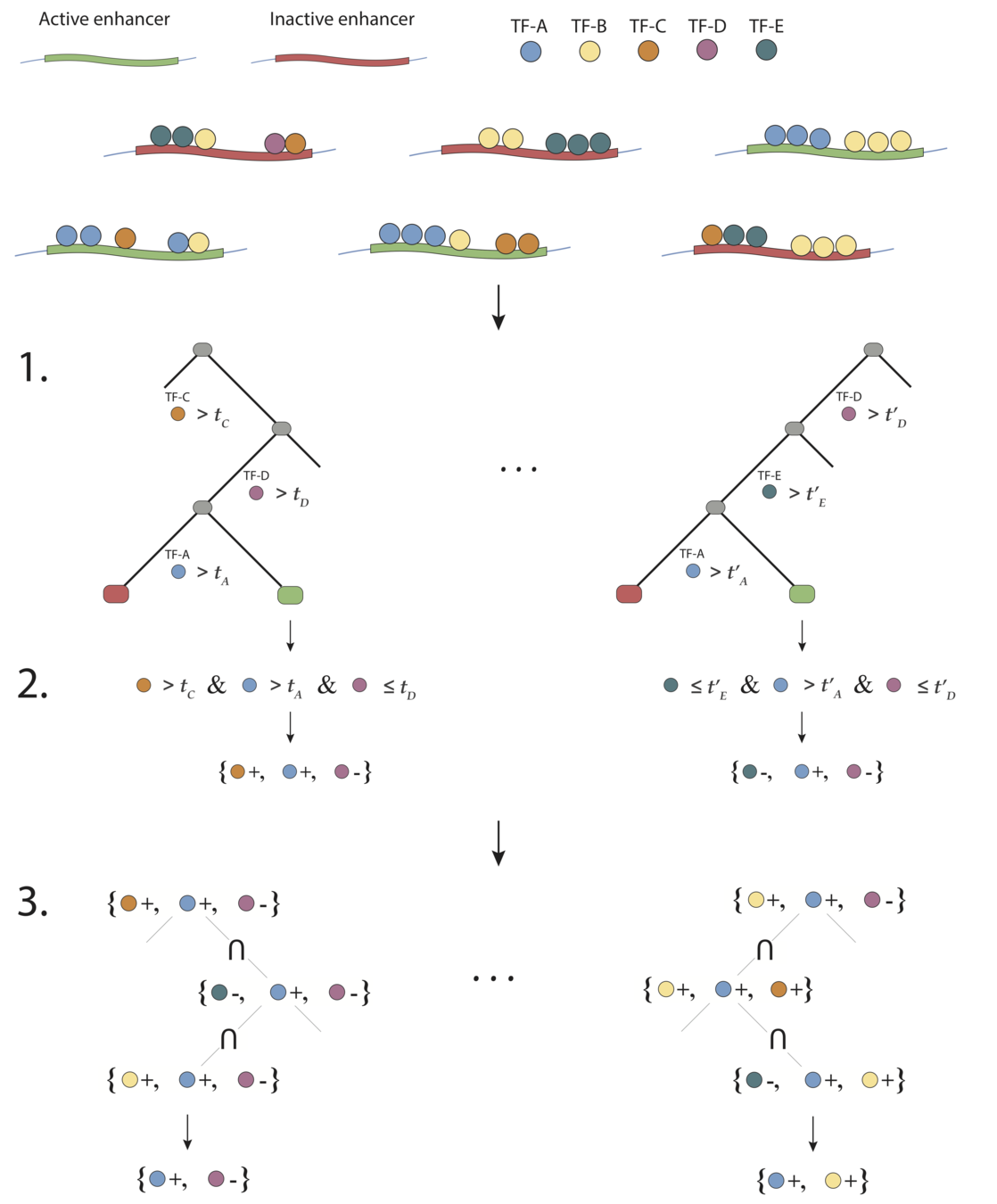
iterative Random Forests (iRF)
&
signed iterative Random Forests (siRF)
Joint work with Sumanta Basu, James B. Brown, Susan Celniker, and Bin Yu
iterative Random Forest to identify high-order interactions in genomic data
- Iteratively re-weighted Random Forests stabilize decision path
- Generalized random intersection trees search for high-order interactions
- Stability bagging evaluates interactions
iterative Random Forests (iRF) build on PCS to identify genomic interactions in developing Drosophila embryos
Open source R implementation: https://cran.r-project.org/web/packages/iRF/
Iteratively re-weighted random forests
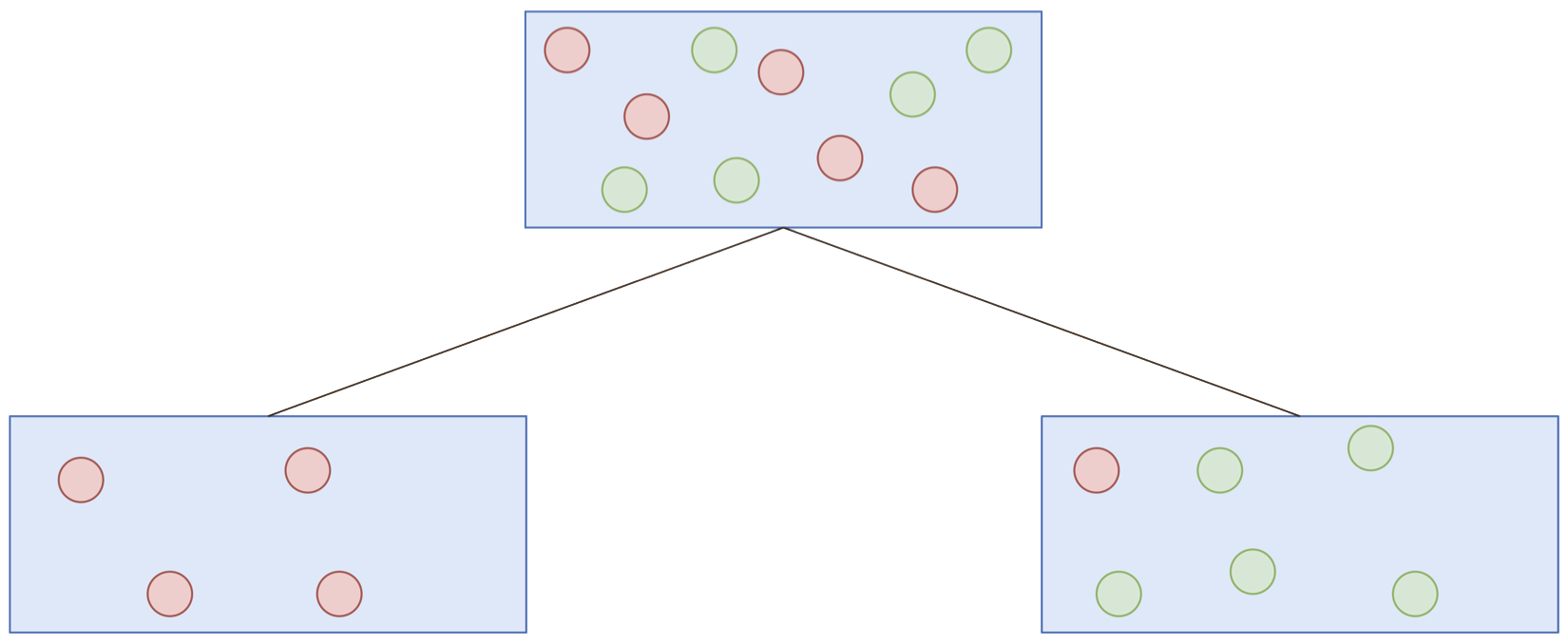
Classification and regression trees (CART)
Breiman et al. (1984)
For current node:
- Select splitting feature and threshold
- Partition data
- Repeat until stopping criteria
Classification and regression trees (CART)
Breiman et al. (1984)

For current node:
- Select splitting feature and threshold
- Partition data
- Repeat until stopping criteria
Random Forests
Breiman (2001)
Random forests modify CART to improve predictive accuracy:
- CART trees are trained on bootstrap samples of the data
- CART criterion evaluated on subset of features sampled uniformly at random
The CART criterion: Gini impurity
Proportion positive responses
Number of observations
Gini impurity:
Decrease in Gini impurity:
Mean decrease in impurity:
On average, how much does splitting on a variable decrease the Gini impurity?
Feature-weighted Random Forests
Amaratunga et al. (2008)
Random forests:
At each node of the decision tree, uniformly sample a subset of features
Feature-weighted random forests:
At each node of the decision tree, sample a subset of features with probability proportional to
Feature weights
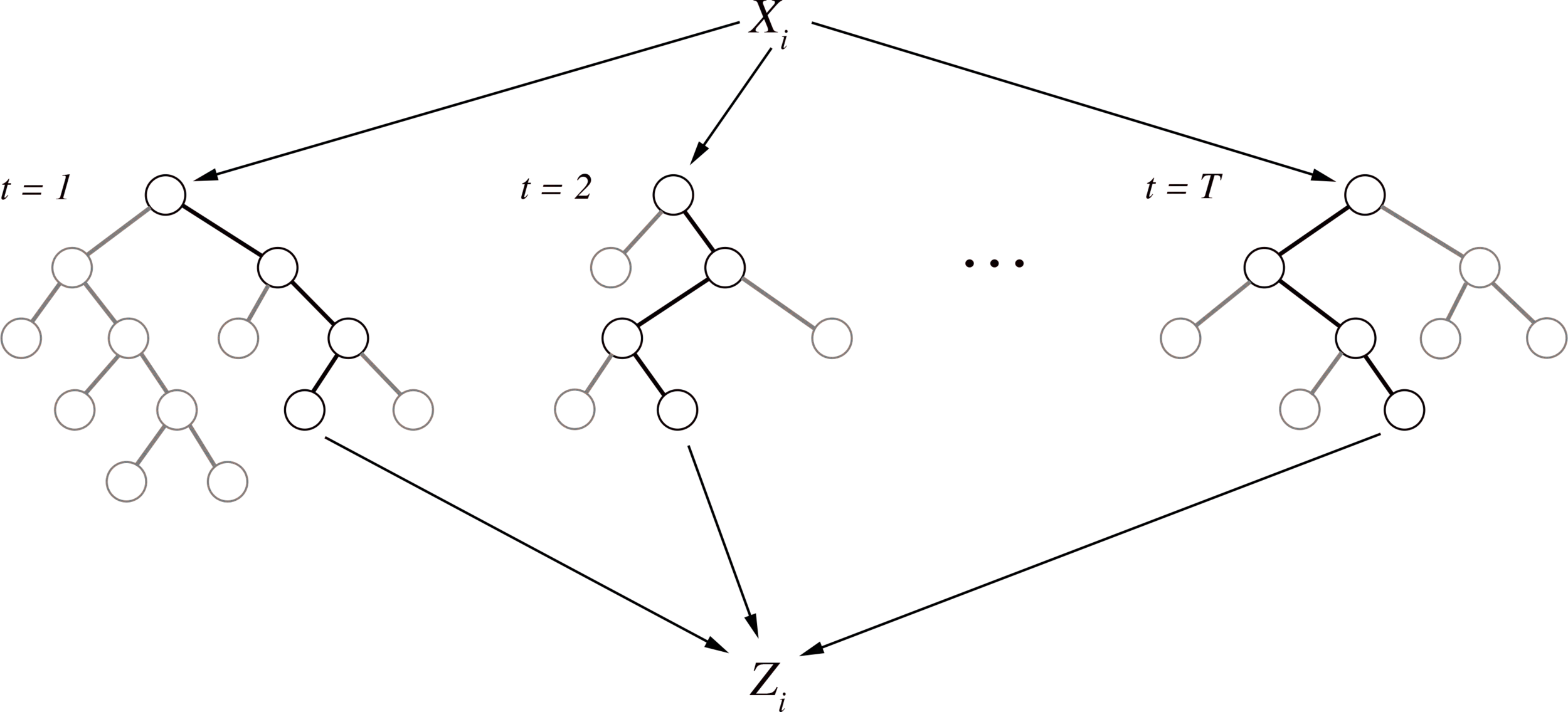
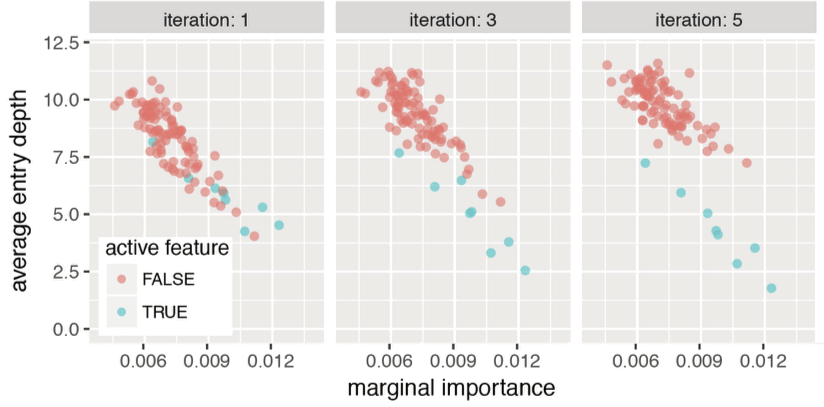
Iterative re-weighting stabilizes random forest decision paths
Gini importance
Iteration 1
Iteration K
Feature weights
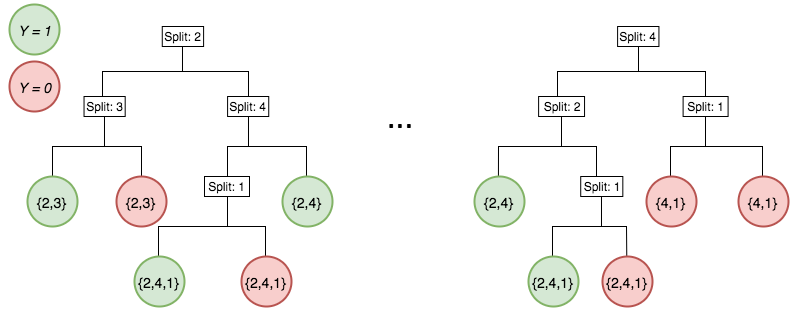
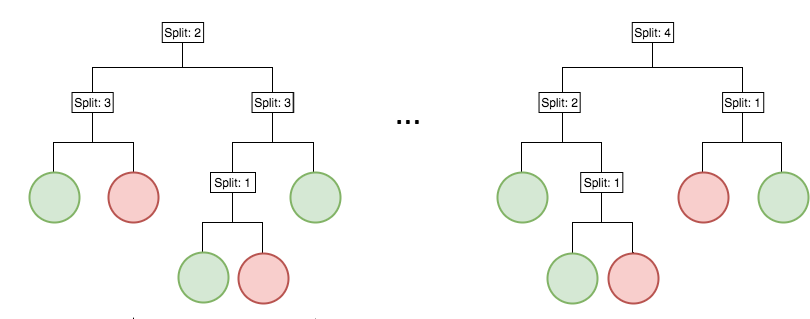
Iterative re-weighting helps recover high-order interactions
Generalized random intersection trees
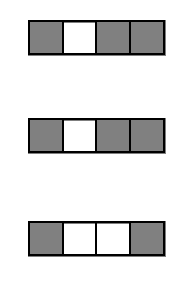
Encoding decision paths to extract active features
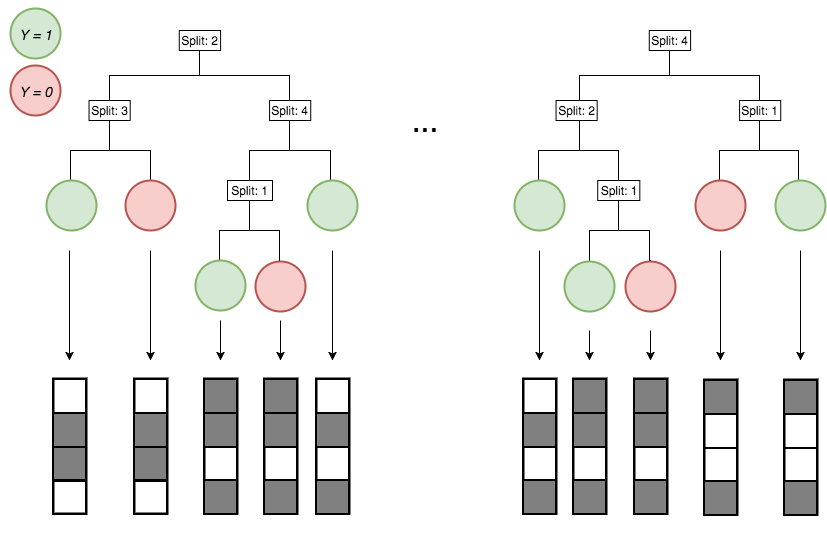
Active
Inactive
Continuous measurements
Binary features
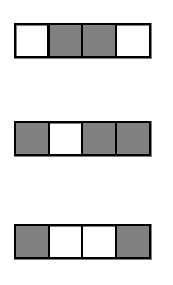
Encoding decision paths to extract enriched and depleted features
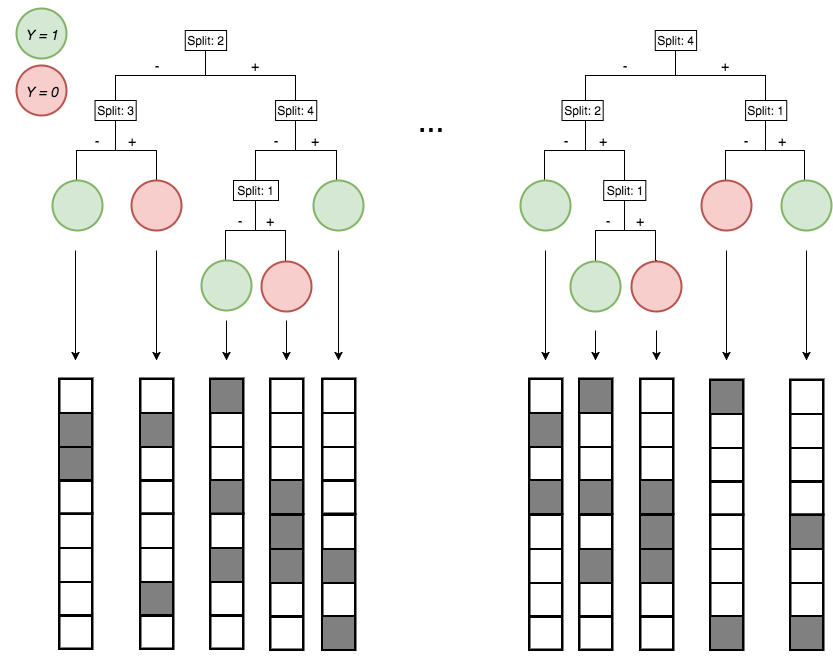
Enriched
Depleted
Generalized random intersection trees search for high-order interactions

1. Iteratively re-weighted random forests
3. RIT on random forest decision paths
2. Decision path feature transformation
.
.
.
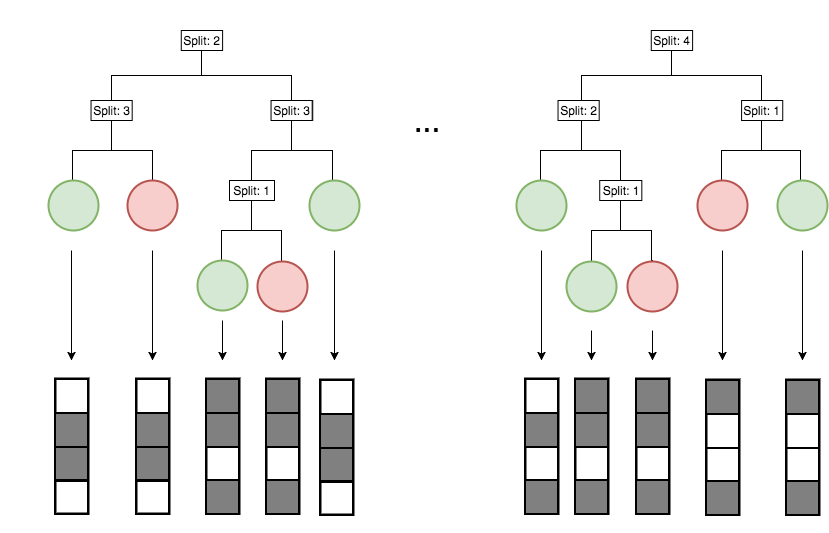
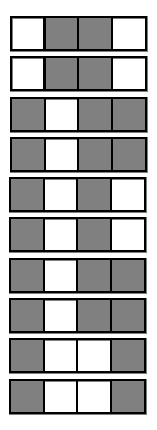
Encoding decision paths to extract prevalent decision rules
Continuous measurements
Binary feature encoding

Decision rules
Encoding decision paths to extract prevalent decision rules
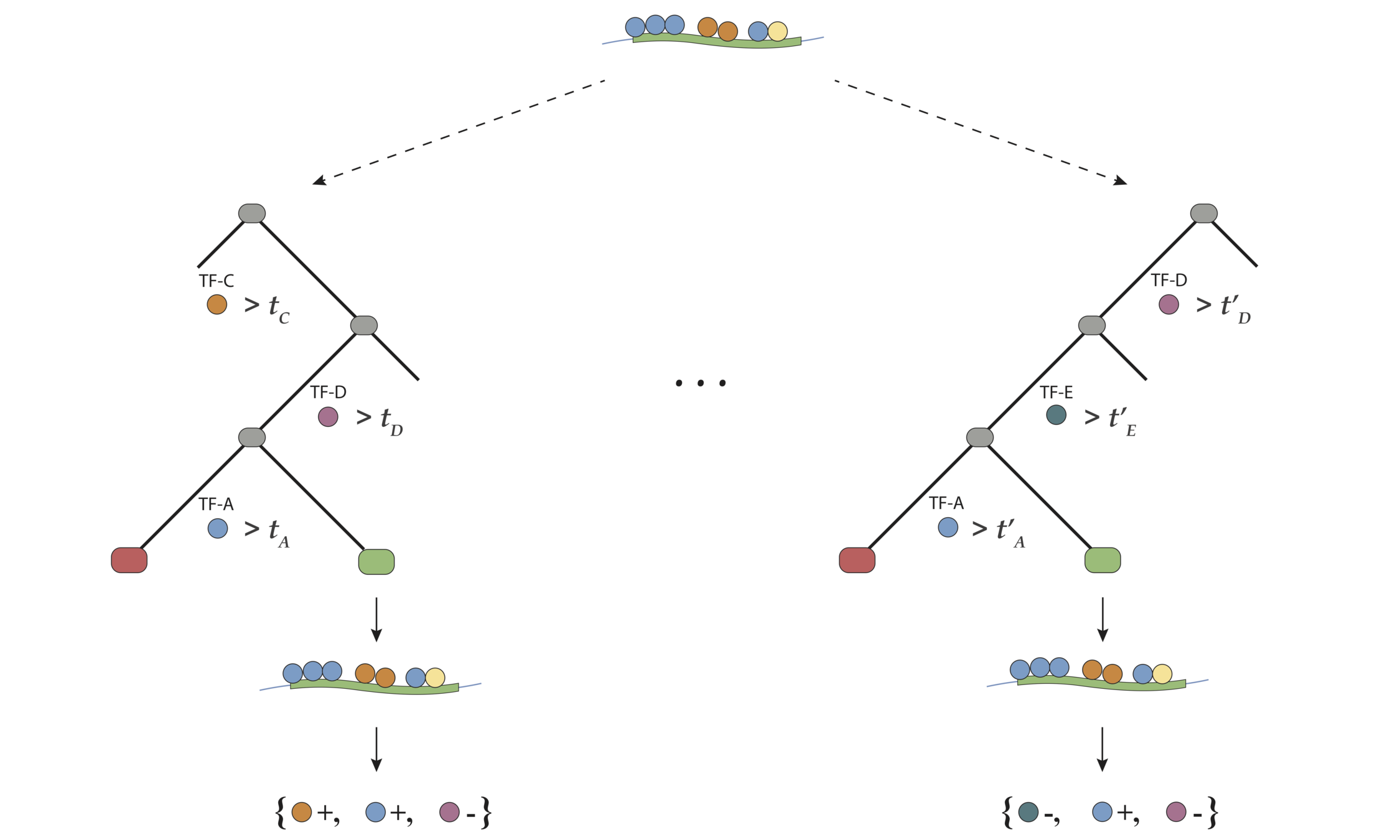
. . .
Generalized random intersection trees search for high-order interactions
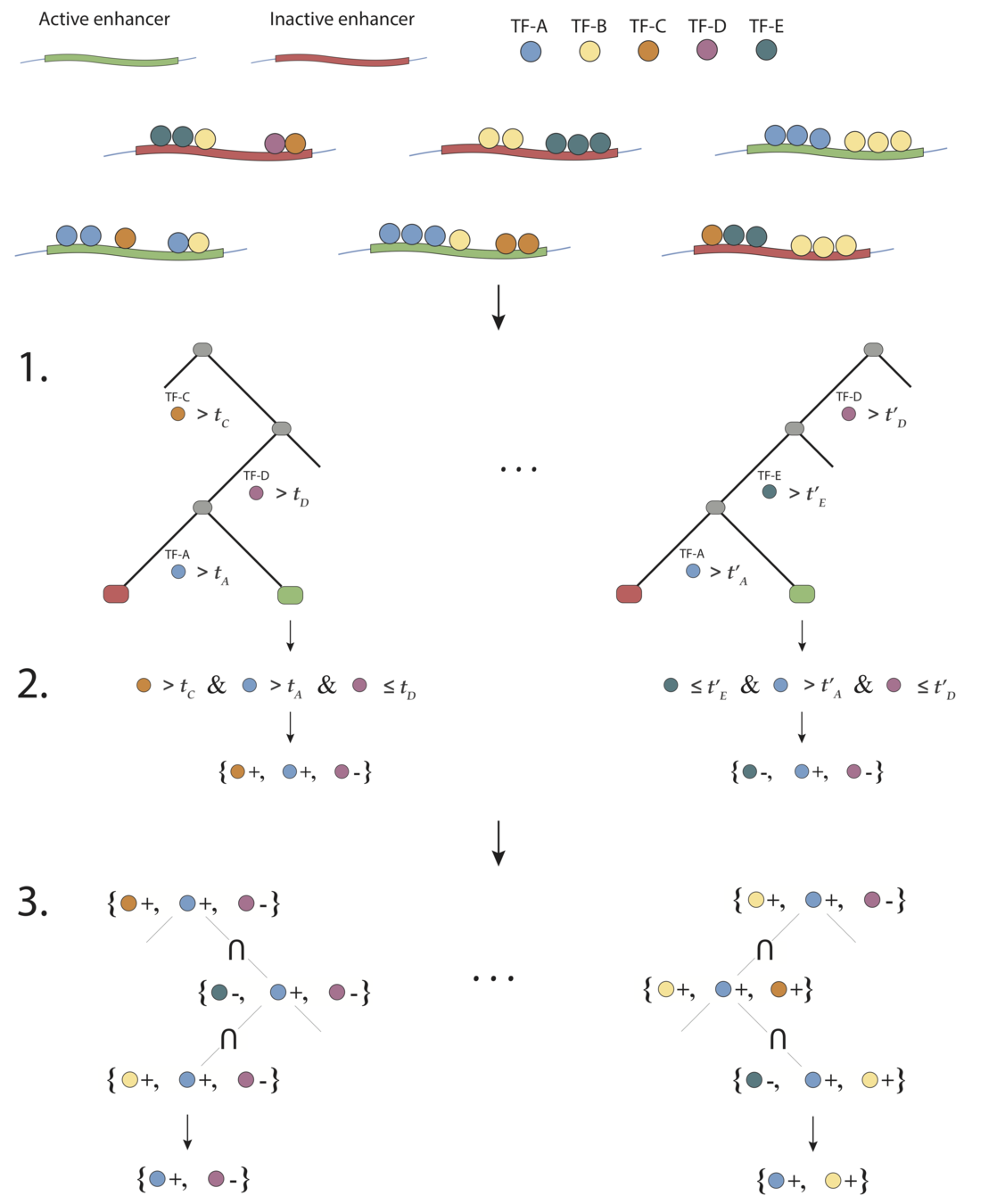
Prevalent interactions
Binary feature encoding
RIT
. . .
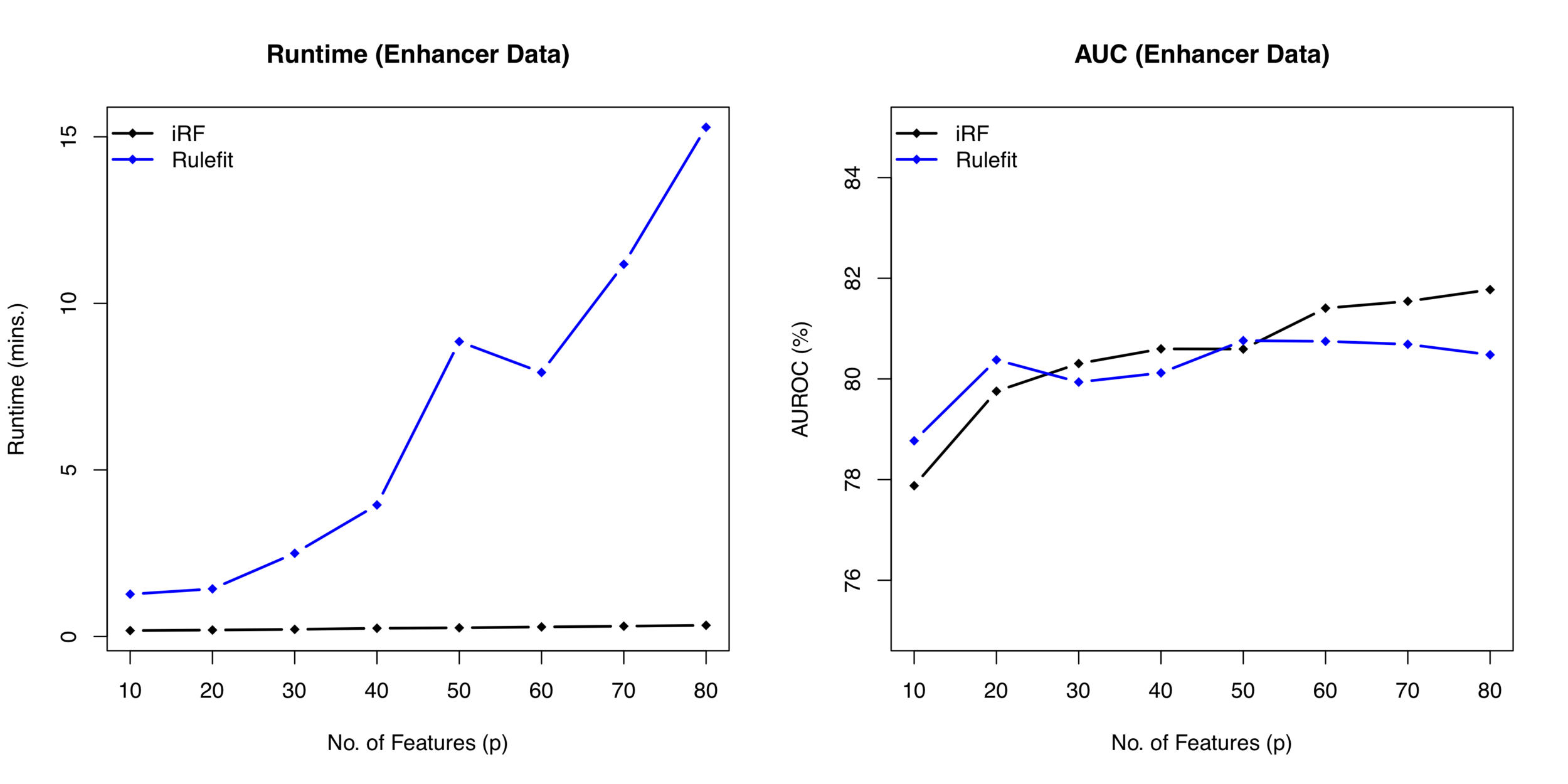
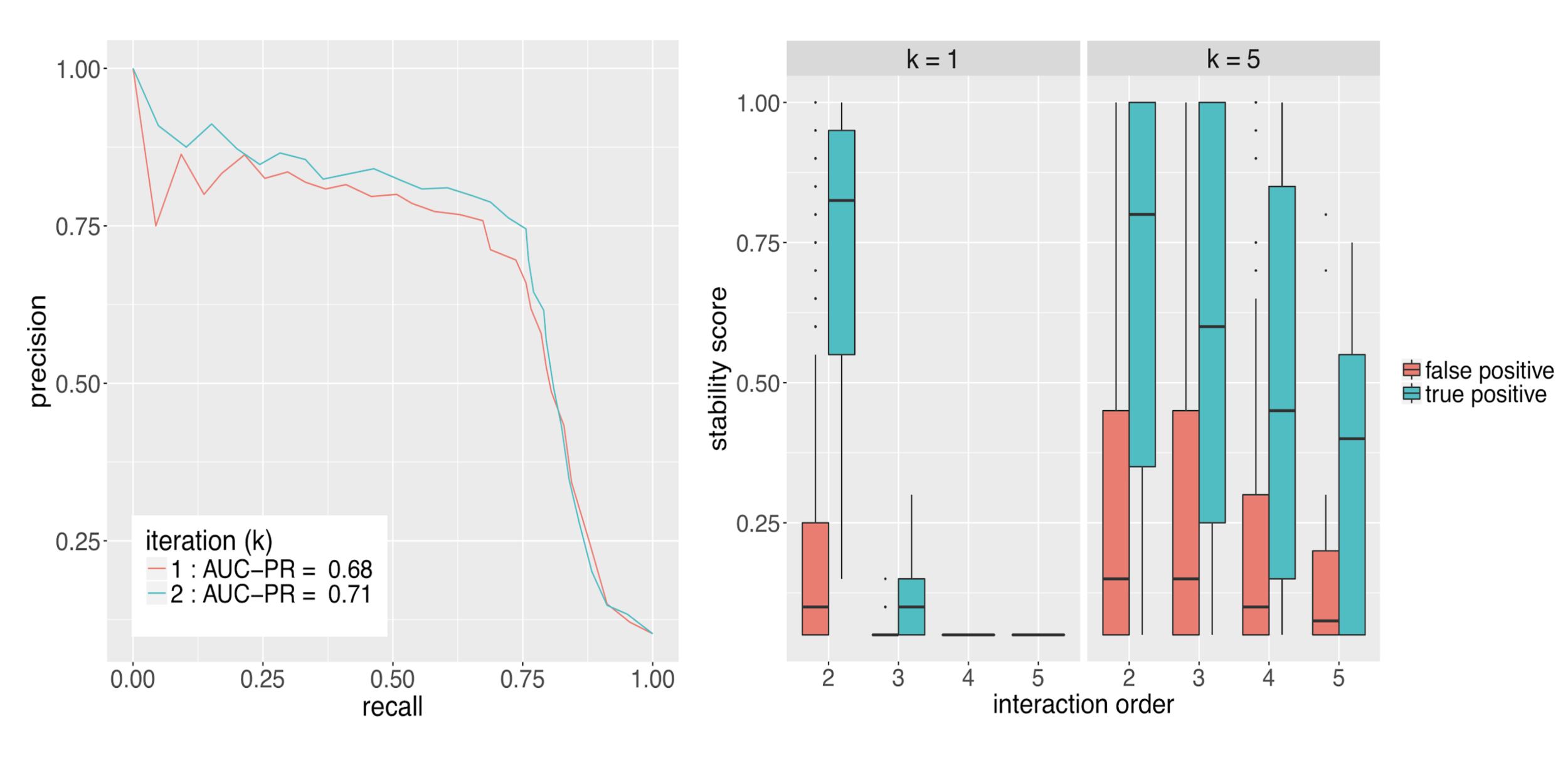
Runtime comparison between iRF and RuleFit

Evaluating interactions
Importance measures for high-order interactions
- Prevalence: how frequently is an interaction observed among positive responses?
- Precision: how accurately does an interaction predict positive responses?
Null metrics describe importance measures under simple structure
- Class precision enrichment (CPE): is an interaction more prevalent among a particular class?
- Feature selection dependence (FSD): are features selected in a dependent manner?
- Mean increase in precision (MIP): do additional features improve prediction accuracy?
Screening interactions based on simple structure (CPE)
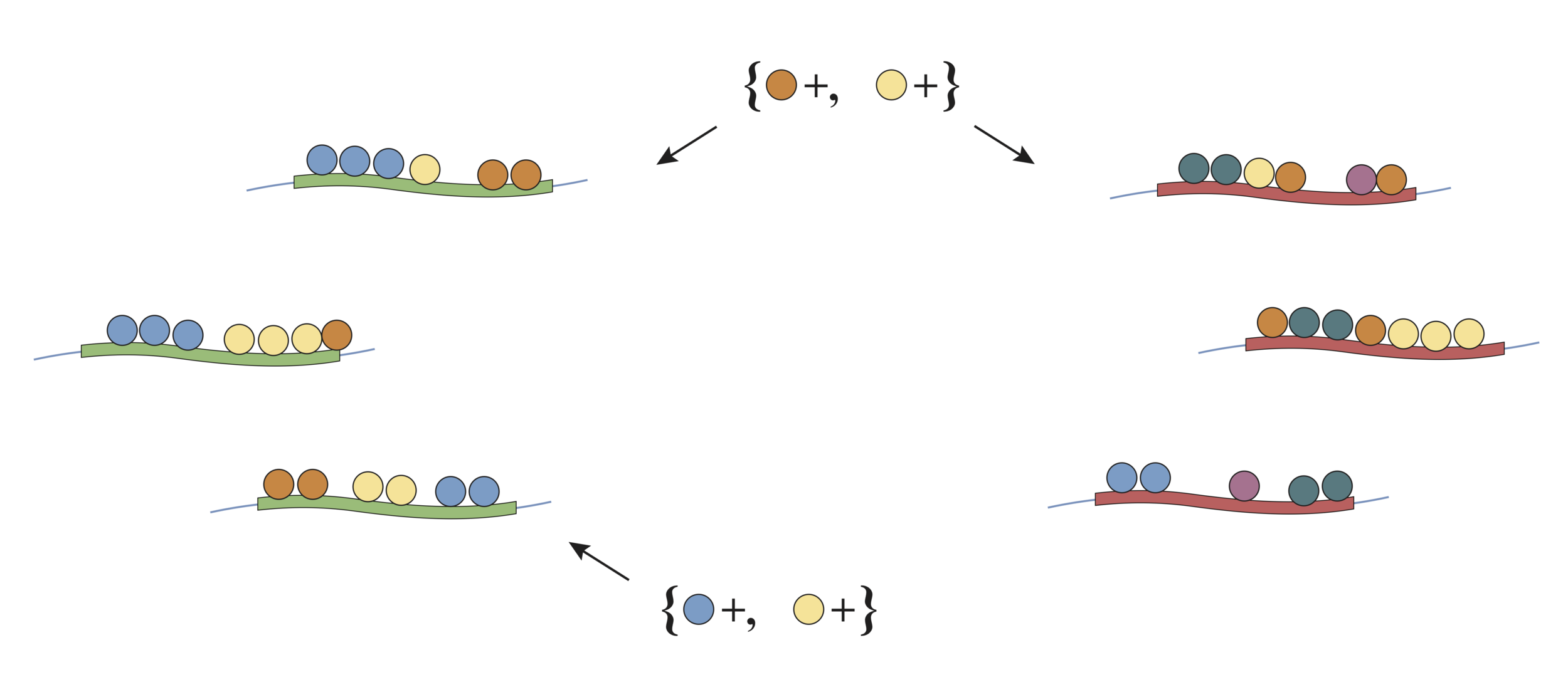
No
Yes
Is an interaction enriched in one class relative to the other?
Prevalence measures the stability of an interaction across an RF
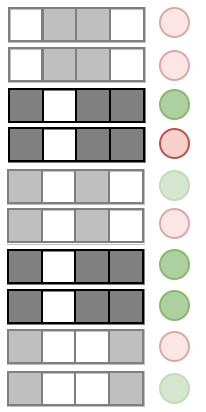
Prevalence:
Examples:
Precision measures the predictive accuracy of an interaction across an RF
Precision:
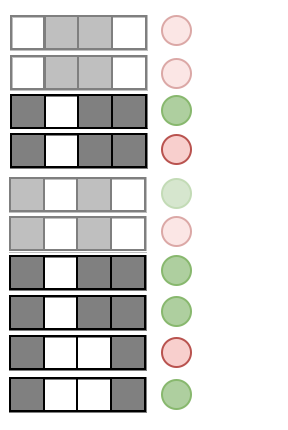
Examples:
Screening interactions based on simple structure (FSD)
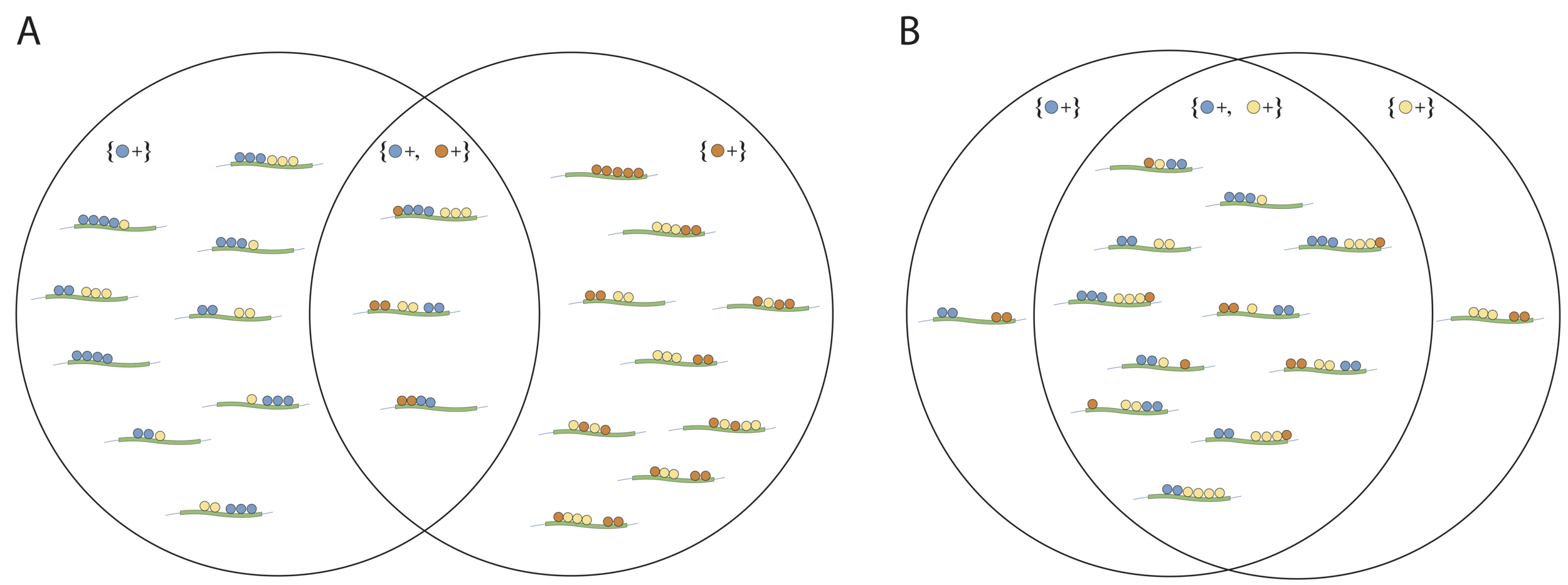
Do features co-occur in a dependent manner?
No
Yes
Screening interactions based on simple structure (MIP)
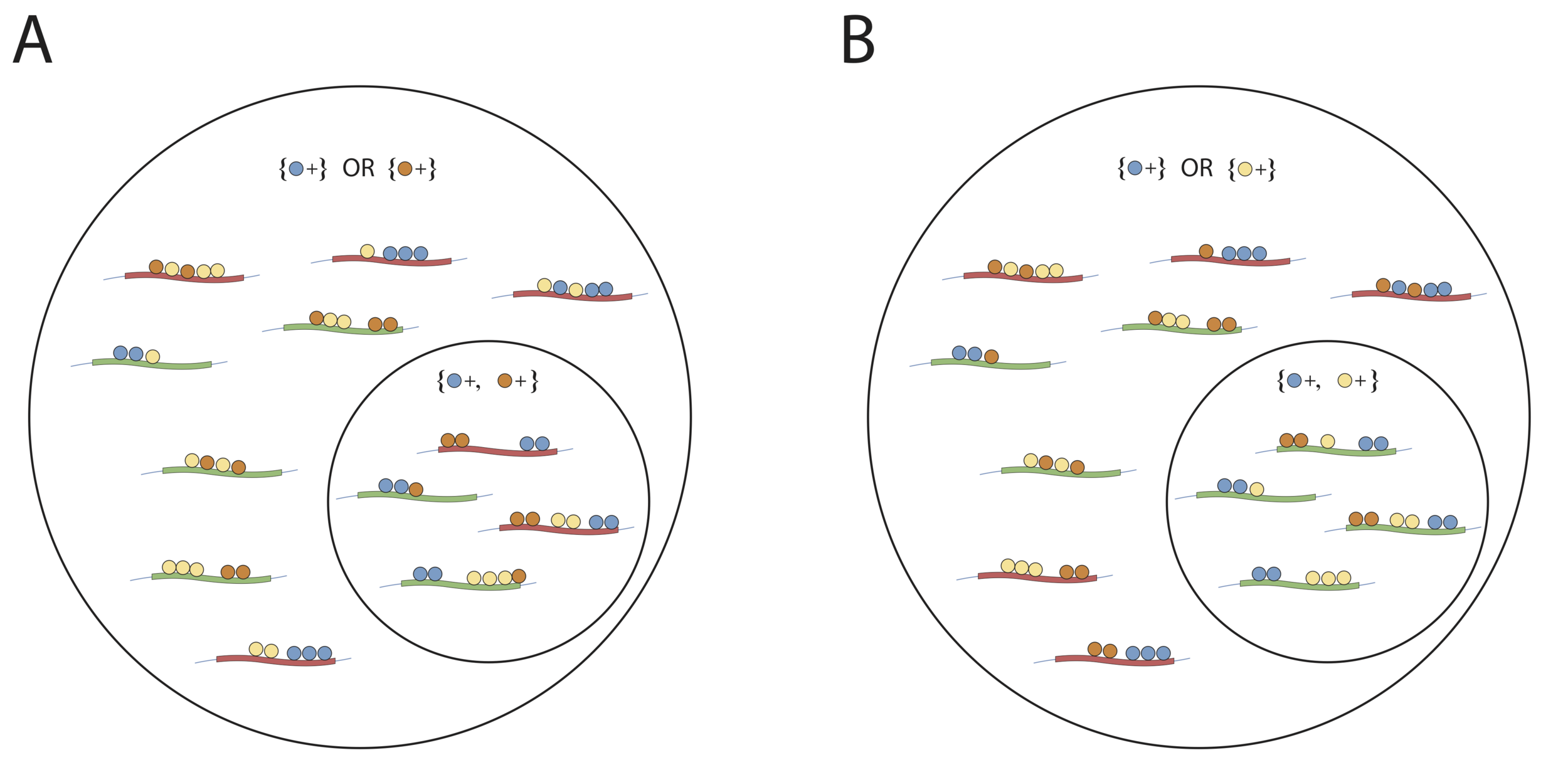
Do additional features allow provide predictive power?
No
Yes
Bagging evaluates stability of interactions across entire iRF workflow relative to resampling








1. Iteratively re-weighted RF stabilize decision paths
2. gRIT searches for high-order interactions along decision paths
3. Importance metrics evaluate interactions in fitted RF
Outer layer bootstrap samples
Case studies in Drosophila
Predicting enhancer activity throughout embryonic development
Enhancers: Pfeiffer et al. 2008, Fisher et al. 2012, Kvon et al. 2014
ChIP: MacArthur et al. 2009, Li et al. 2008, modENCODE/modERN consortia
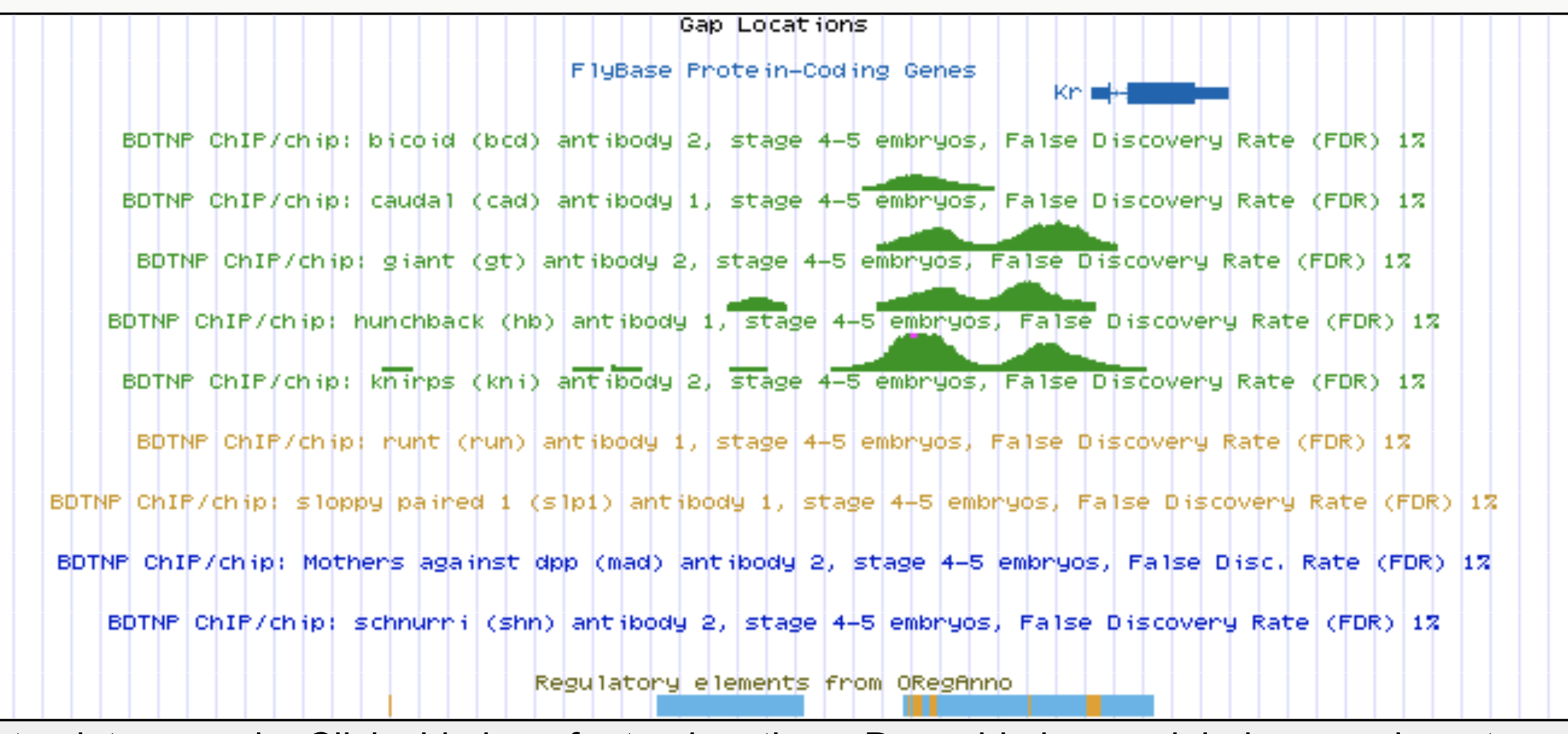
Predicting enhancer activity throughout embryonic development
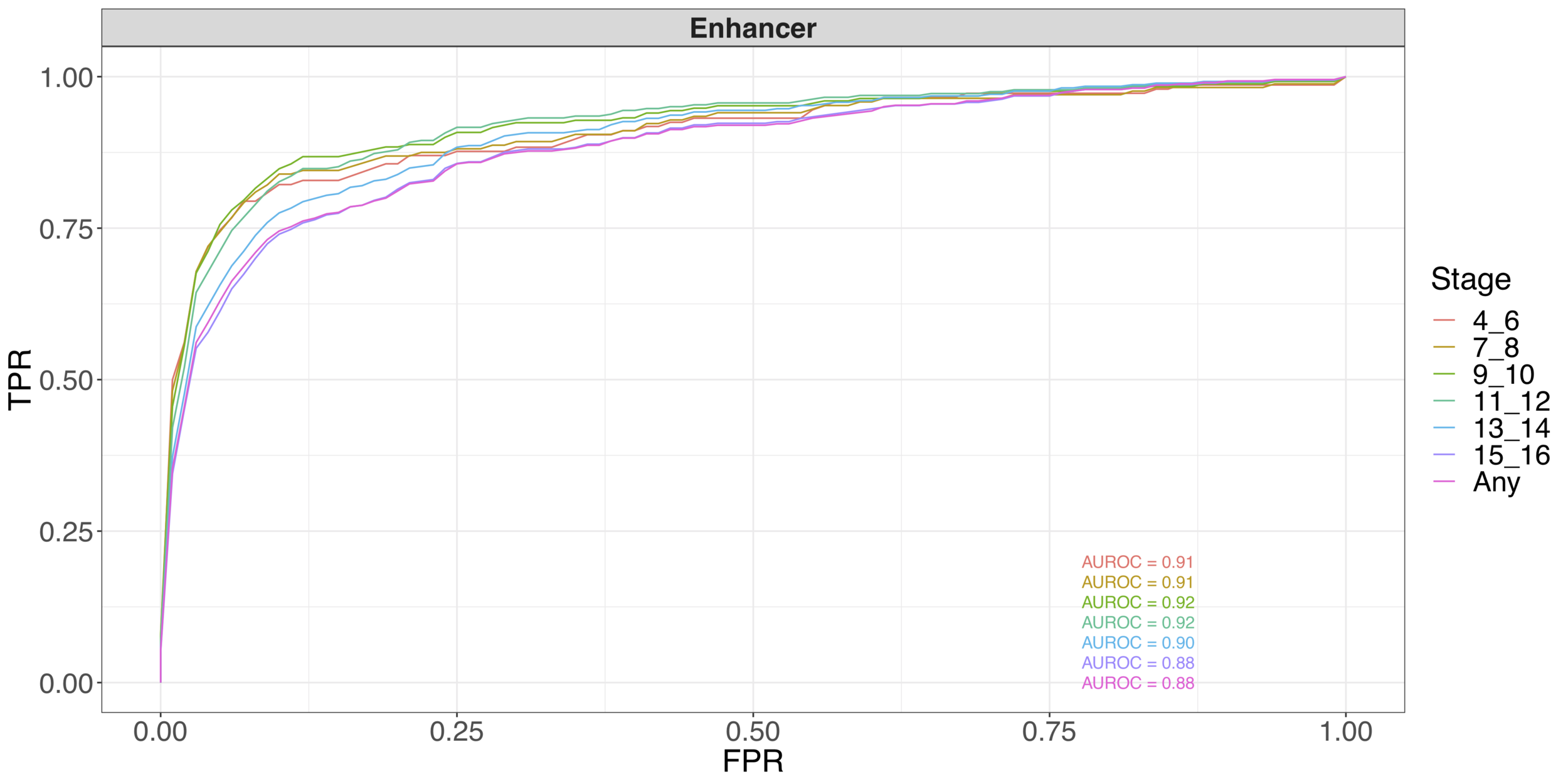
Early stage (not shown): 24 TFs; Basu, K., Brown, and Yu (2018)
All stages (shown): 307 TFs; K., Basu, Brown, Celniker, and Yu (2019)
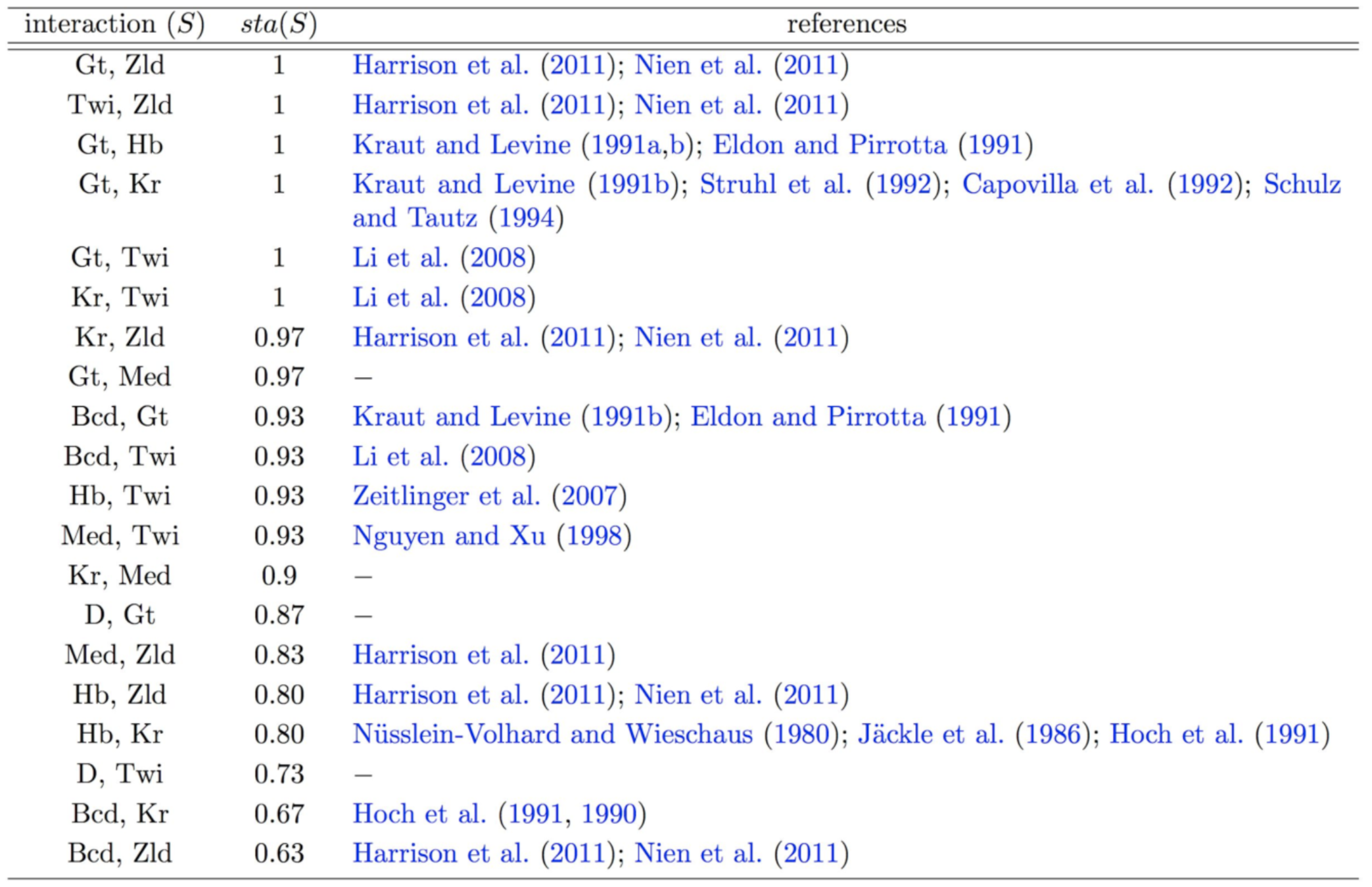
iterative Random Forests recover well-known pairwise interactions
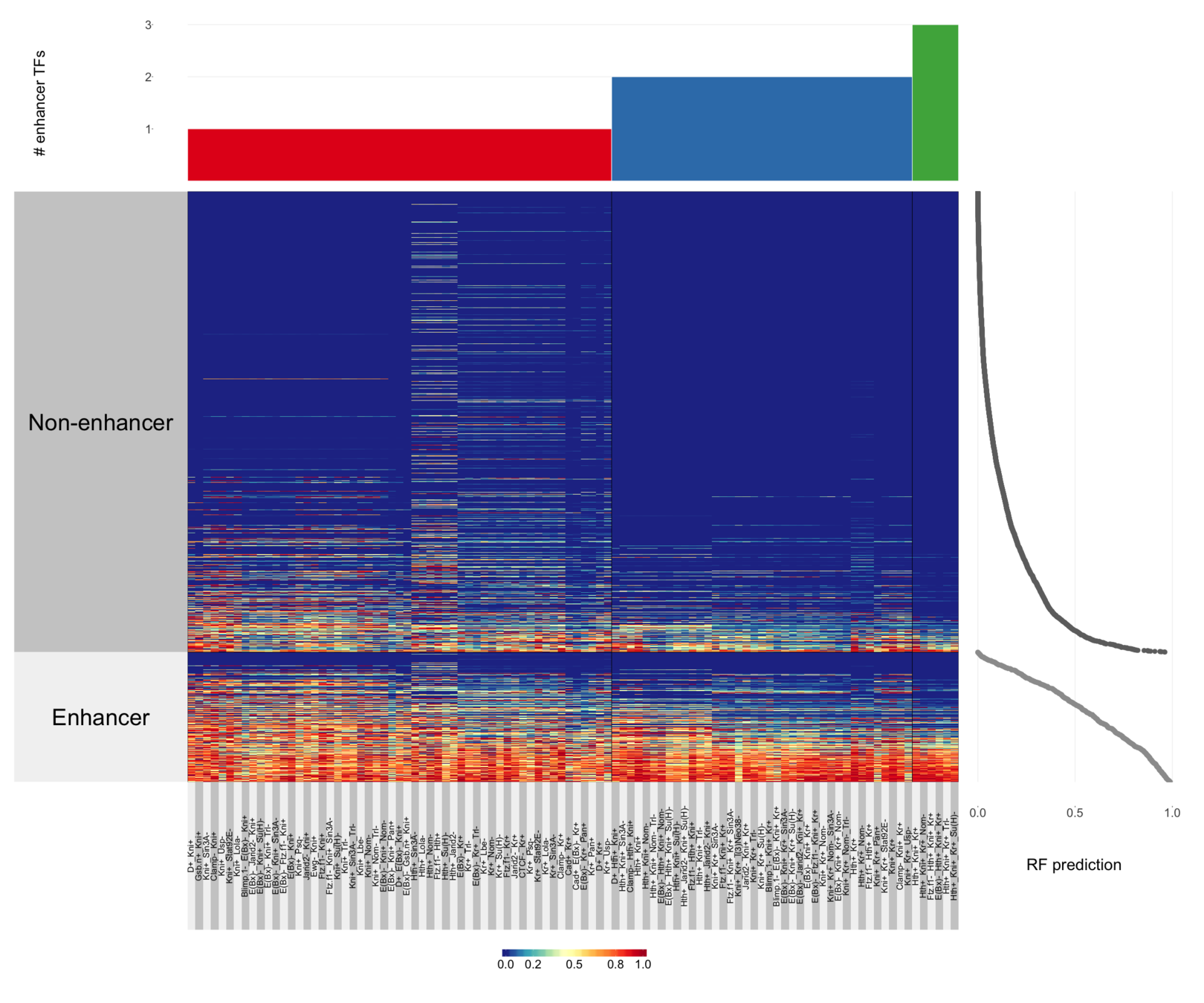
Signed interactions define active enhancers with high precision
RF-prediction
0
1
Interaction-based predictions
Signed interactions define active enhancers with high precision

RF-prediction
0
1
Interaction-based predictions
High false
negative rate
Missing
data (stage)
TFs involved in important interactions are measured at "high confidence" stages

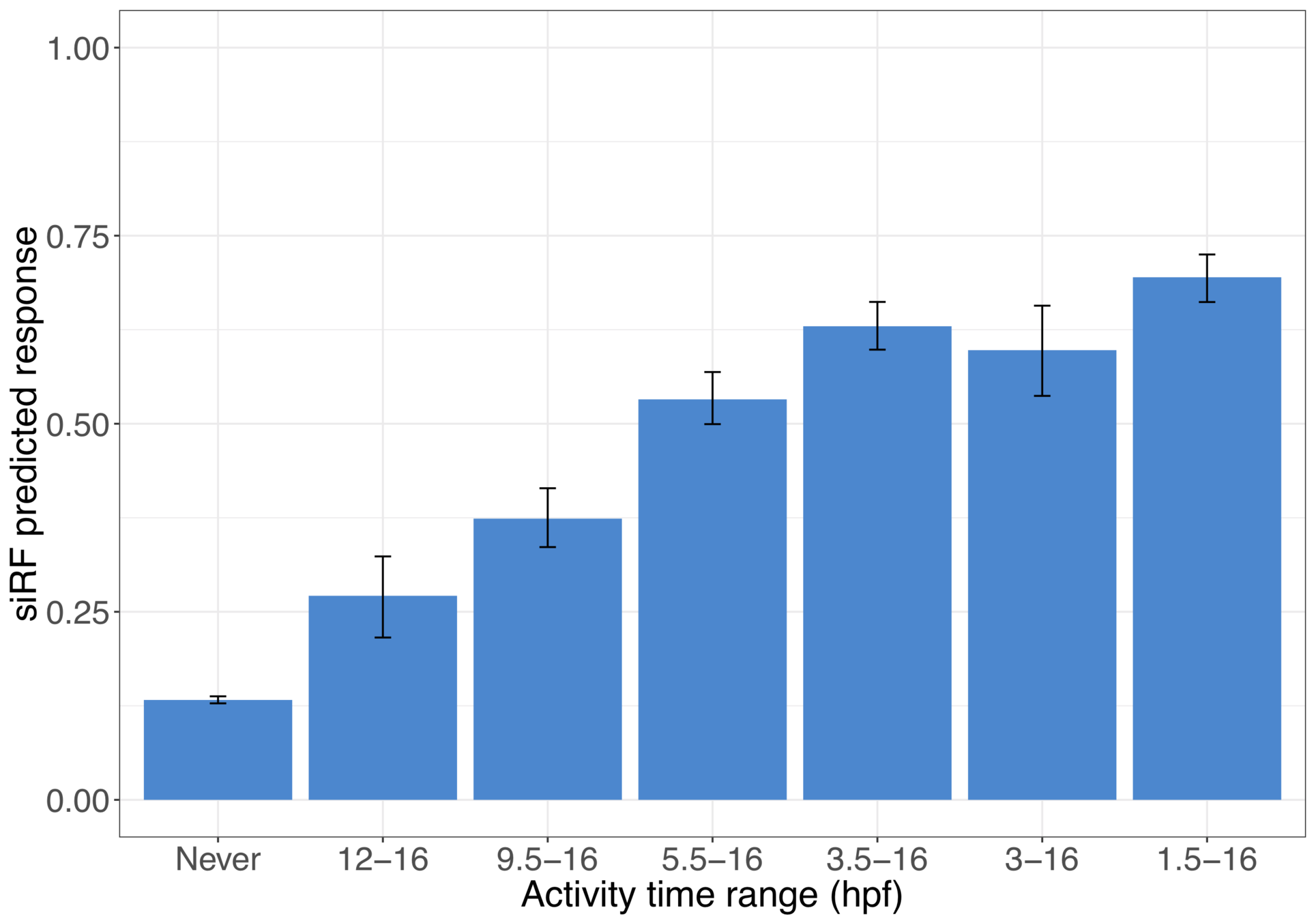
Activated late
Activated early
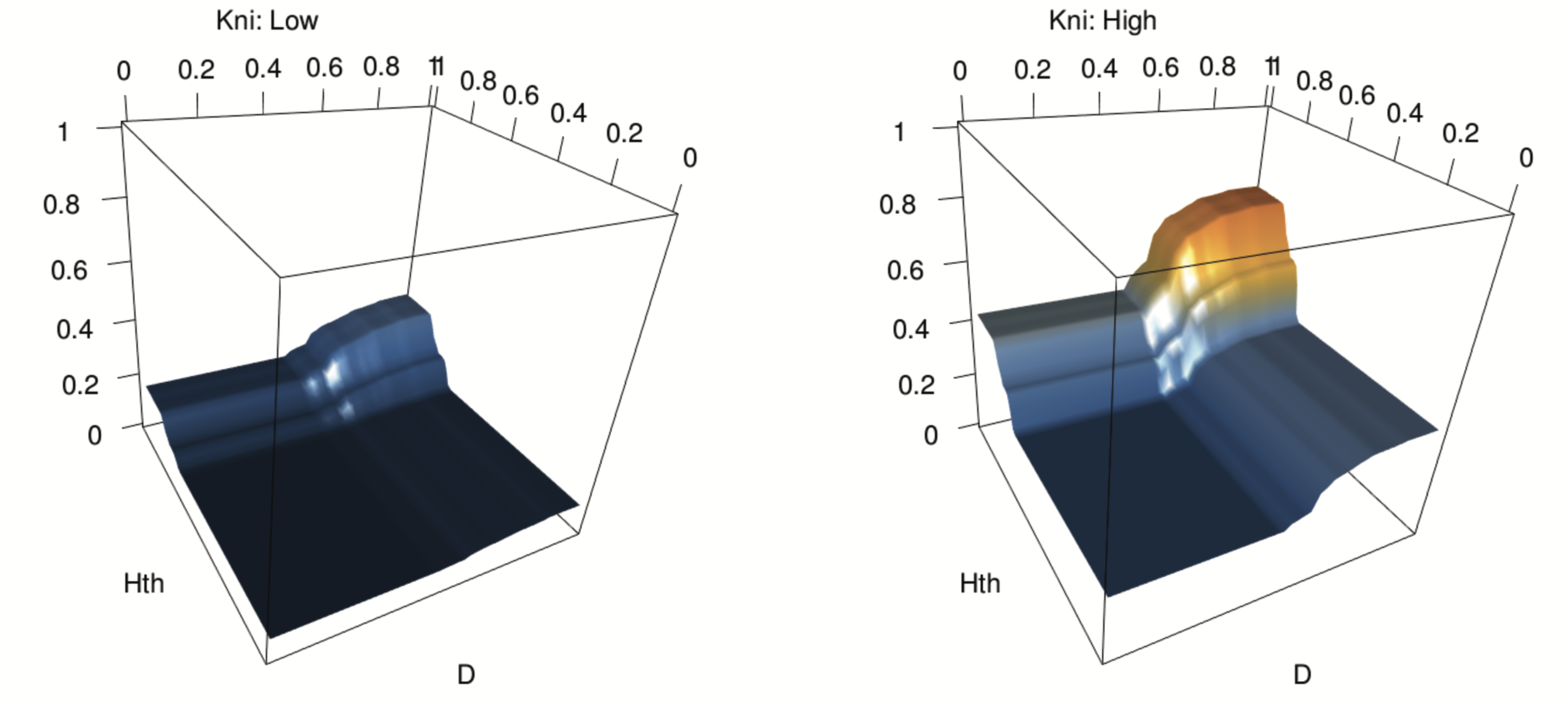
Novel order-3 interactions exhibit AND-like behavior (D, Hth, Kni)
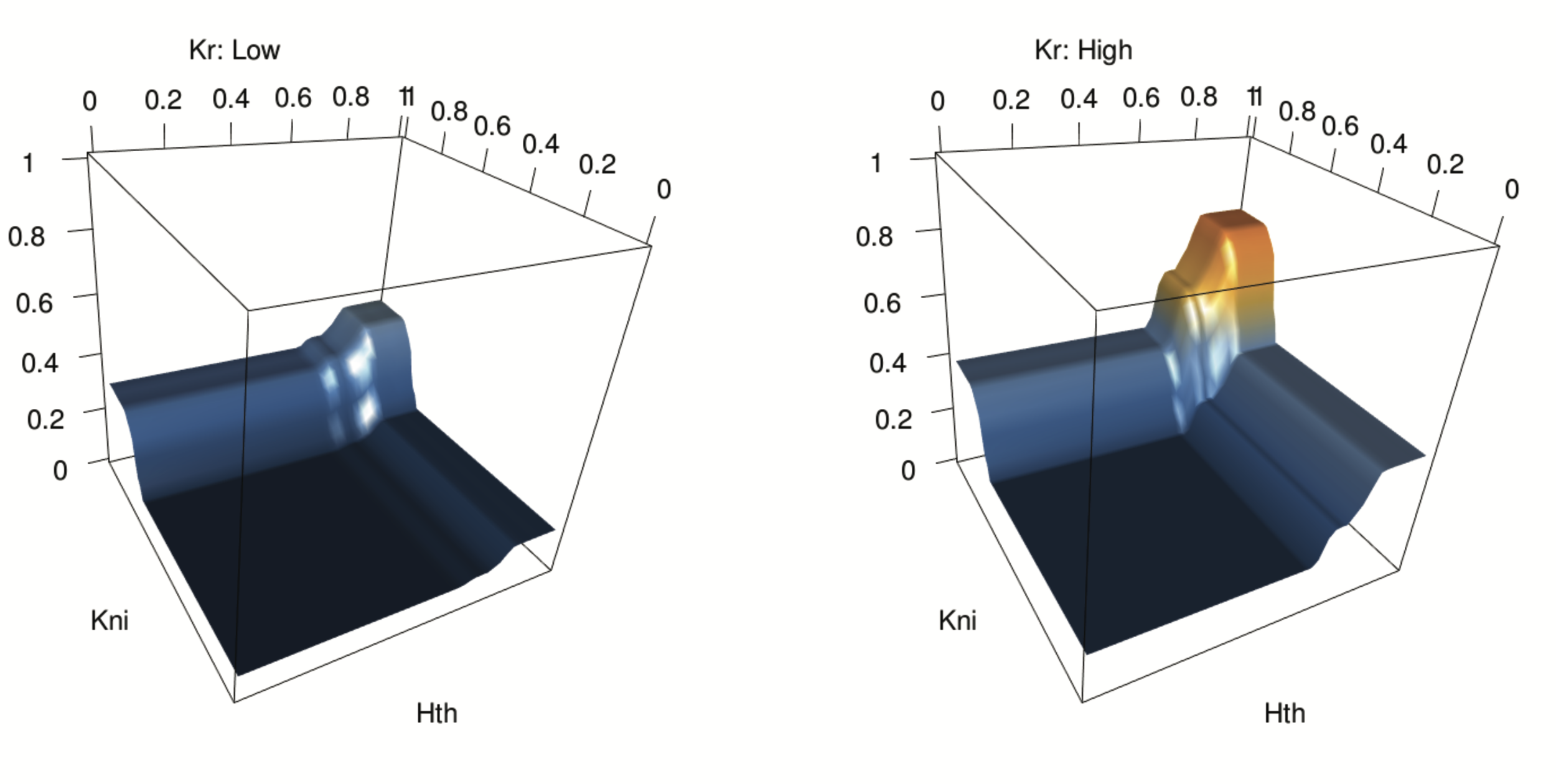
Novel order-3 interactions exhibit AND-like behavior (Hth, Kni, Kr)
Putative interactions define targets for follow-up experiments

High confidence enhancer elements
Proposed mechanism: Hth+ & Kni+ & Kr+
Summary
- iRF and siRF identify well known interactions in Drosophila and posit new, high-order interactions surrounding important regulatory factors in the early embryo.
- By decoupling interaction order from the computational cost of discovery, iRF and siRF allow us to investigate mechanisms in genome biology and beyond.
Ackowledgements




S. Basu
J. Brown
B. Yu
S. Celniker
E. Frise






Yu Group
Thank You!
siRF
By kkumbier
siRF
- 71



