

\text{Weekly Chat}
\text{March 2, 2021}
\text{Fourier Shape Descriptor}
\text{SORT}
\text{3d Visualization}
\text{Graph Matching}
\text{MitoEM}
\text{Fourier Shape Descriptor}
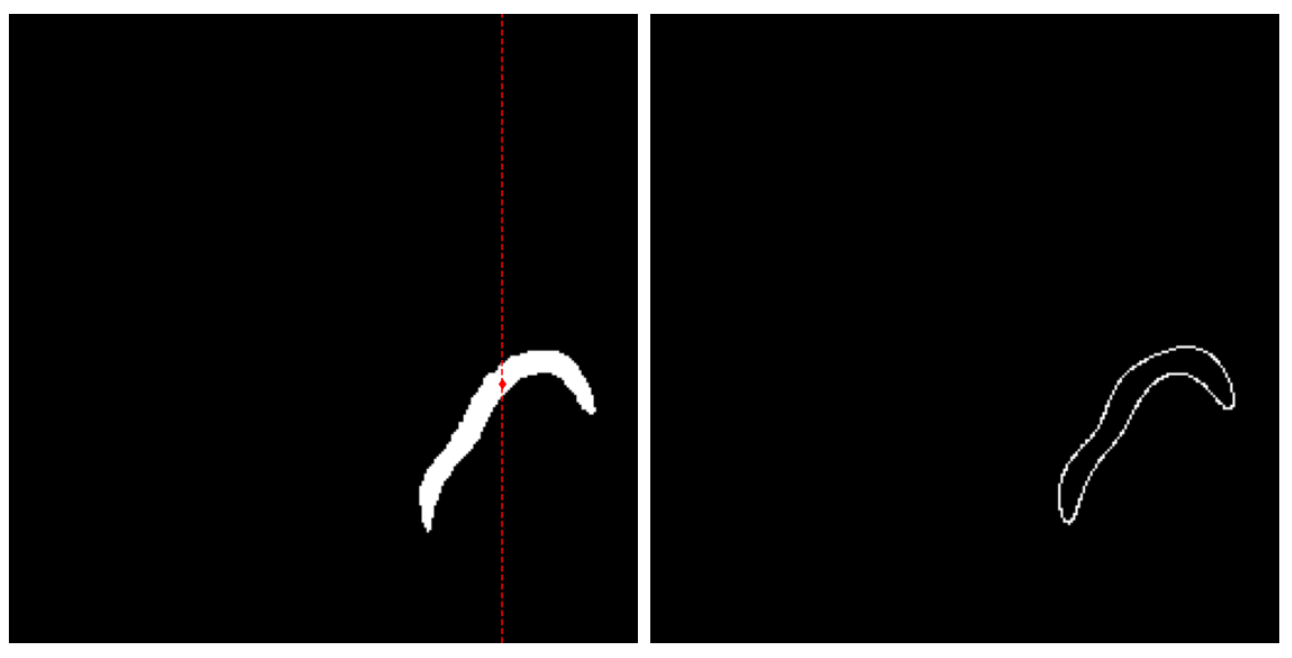
\text{Fourier Shape Descriptor}
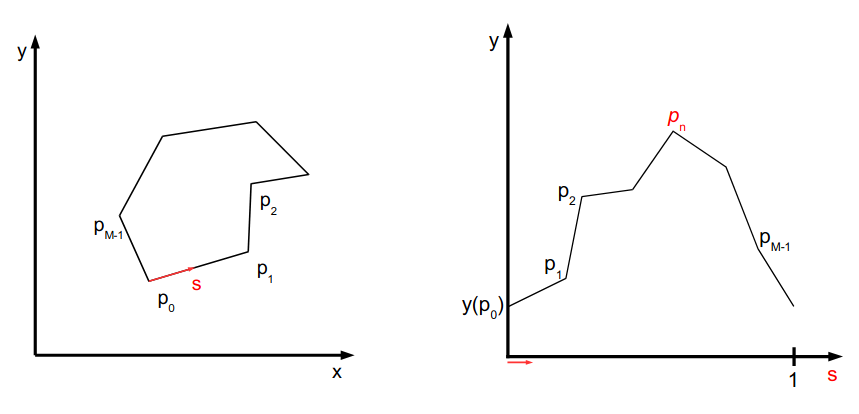
\text{Fourier Shape Descriptor - idea is ...}
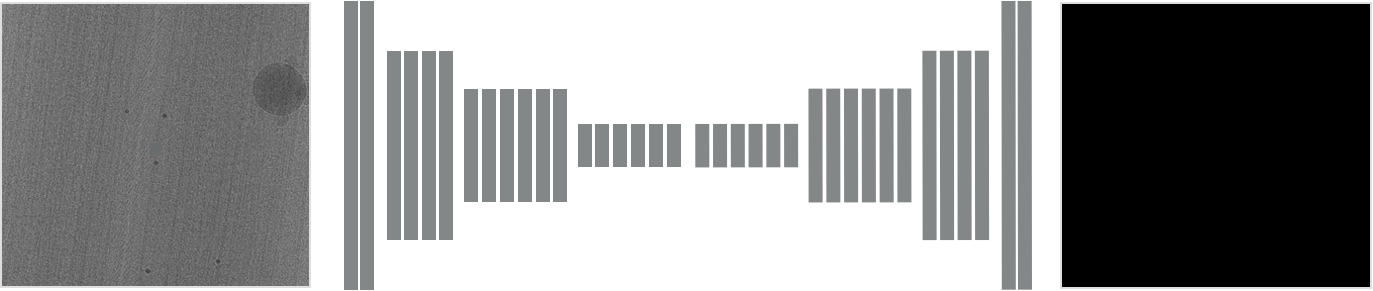
6 \times 4 \text{ predictions p.p}
\text{Fourier Shape Descriptor - idea is ...}

6 \times 4 \text{ predictions p.p}
\text{IDFT}
\text{x \& y locations in cartesian space}
\text{Fourier Shape Descriptor - idea is ...}

6 \times 4 \text{ predictions p.p}
\text{IDFT}
\text{x \& y locations in cartesian space}
\text{differentiable IOU loss}
\text{Fourier Shape Descriptor - idea is ...}

6 \times 4 \text{ predictions p.p}
\text{IDFT}
\text{x \& y locations in cartesian space}
\text{differentiable IOU loss}
\text{translation invariance loss}
\text{Fourier Shape Descriptor - idea is ...}

6 \times 4 \text{ predictions p.p}
\text{IDFT}
\text{x \& y locations in cartesian space}
\text{differentiable IOU loss}
\text{translation invariance loss}
\text{foreground loss}
\text{Fourier Shape Descriptor - idea is end-to-end training}

6 \times 4 \text{ predictions p.p}
\text{IDFT}
\text{x \& y locations in cartesian space}
\text{differentiable IOU loss}
\text{translation invariance loss}
\text{foreground loss}
\text{Fourier Shape Descriptor - questions}
\text{How to compute interior area of points defining contour in differentiable manner?}
\text{Are there unique solutions which the network can achieve during training?}
\text{Fourier Shape Descriptor}
\text{SORT}
\text{3d Visualization}
\text{Graph Matching}
\text{MitoEM}
\text{SORT}
\text{T=0}
\text{50}
\text{100}
\text{150}
\text{200}
\text{250}
\text{350}
\text{300}
\text{600}
\text{SORT}
\text{Scheme Three}
\text{T=0}
\text{T=1}
\text{T=2}
\text{Hard Assignment}
\text{Real Time}
\text{Local Optima}
\text{Correct for mitosis - conservation of mass!}




\text{...}




\text{...}
p^{*}_{1}
p^{*}_{2}
p^{*}_{3}
p^{*}_{4}
p^{*}_{...}
p^{*}_{M}
q_{2}
q_{3}
q_{4}
q_{N}
q_{...}
\text{4.1}
\text{2.6}
\text{11.3}
\text{89.7}
\text{14.2}
\text{90.8}
\text{64.2}
\text{7.8}
\text{16.9}
\text{70.8}
\text{16.7}
\text{32.4}
\text{9.7}
\text{15.4}
\text{8.3}
\text{20.1}
\text{11.6}
\text{14.9}
\text{18.1}
\text{3.7}
\text{24.1}
\text{36.4}
\text{12.2}
\text{9.5}
\text{14.2}
\text{17.5}
\text{23.45}
\text{6.7}
\text{19.9}
\text{22.1}
\text{14.8}
\text{16.4}
\text{63.7}
\text{21.5}
\text{81.1}
\text{9.6}
q_{1}
\textit{to obtain one-to-one matching }
C_{ij} := C(p^{*}_{i}, q_{j})
\hat{X}= \underset{X}{\text{arg min}} \sum_{i=1}^{M} \sum_{j=1}^{N} C_{ij} X_{ij}
\text{ where } X_{ij} \in \{0, 1\}
\text{ s.t.} \sum_{k=1}^{k=M} 1 \leq X_{ik} \leq 2
\sum_{k=1}^{k=N} X_{kj} = 1
\textit{Apply Hungarian Algorithm}
\text{IOU btw predicted kalman states and actual target states}
\text{Correct for mitosis - conservation of mass!}




\text{...}




\text{...}
p^{*}_{1}
p^{*}_{2}
p^{*}_{3}
p^{*}_{4}
p^{*}_{...}
p^{*}_{M}
q_{2}
q_{3}
q_{4}
q_{N}
q_{...}
\text{4.1}
\text{2.6}
\text{11.3}
\text{89.7}
\text{14.2}
\text{90.8}
\text{64.2}
\text{7.8}
\text{16.9}
\text{70.8}
\text{16.7}
\text{32.4}
\text{9.7}
\text{15.4}
\text{8.3}
\text{20.1}
\text{11.6}
\text{14.9}
\text{18.1}
\text{3.7}
\text{24.1}
\text{36.4}
\text{12.2}
\text{9.5}
\text{14.2}
\text{17.5}
\text{23.45}
\text{6.7}
\text{19.9}
\text{22.1}
\text{14.8}
\text{16.4}
\text{63.7}
\text{21.5}
\text{81.1}
\text{9.6}
q_{1}
\textit{to obtain one-to-one matching }
C_{ij} := C(p^{*}_{i}, q_{j})
\hat{X}= \underset{X}{\text{arg min}} \sum_{i=1}^{M} \sum_{j=1}^{N} C_{ij} X_{ij}
\text{ where } X_{ij} \in \{0, 1\}
\text{ s.t.} \sum_{k=1}^{k=M} 1 \leq X_{ik} \leq 2
\sum_{k=1}^{k=N} X_{kj} = 1
\textit{Apply Hungarian Algorithm}
\text{IOU btw predicted kalman states and actual target states}
\text{Future: incl apppearance vector from EmbedSeg}
\text{Fourier Shape Descriptor}
\text{SORT}
\text{3d Visualization}
\text{Graph Matching}
\text{MitoEM}
\text{Graph Matching}
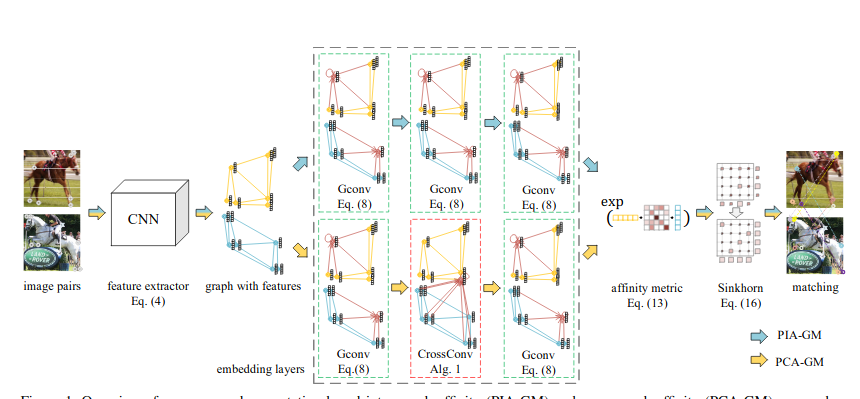
\text{Graph Matching}

\text{Jointly train EmbedSeg on multi modalities}
\text{Same 12 landmarks are annotated in fixed and live embryo}
\text{which means during training we can sample either two fixed embryos}
\text{or two live embryos}
\text{or a fixed and a live embryo}
\text{or the complete set of GT matches using fixed and simulated live}
\text{Graph Matching}

\text{Jointly train EmbedSeg on multi modalities}
\text{Same 12 landmarks are annotated in fixed and live embryo}
\text{which means during training we can sample either two fixed embryos}
\text{or two live embryos}
\text{or a fixed and a live embryo}
\text{or the complete set of GT matches using fixed and simulated live}
\text{currently trying to consolidate and extend these 12 landmark pairs}
\text{Fourier Shape Descriptor}
\text{SORT}
\text{3d Visualization}
\text{Graph Matching}
\text{MitoEM}
\text{3d Visualization}
\text{Create VTK models for all objects}
\text{Figure out how to not introduce rendering artifacts in gray-scale image}
\text{Provide link on project page for users to interact}
\text{Fourier Shape Descriptor}
\text{SORT}
\text{3d Visualization}
\text{Graph Matching}
\text{MitoEM}
\text{MitoEM}
\text{1024}
\text{1024}
\text{MitoEM}
\text{MitoEM}
+
+
*
*
\text{aff(}
+
+
\text{)}

\text{MitoEM}
+
+
*
\text{aff(}
+
+
\text{)}

+
+
*
*
\text{aff(}
+
\text{)}

+
\text{MitoEM}
+
+
*
\text{aff(}
+
+
\text{)}

+
+
*
*
\text{aff(}
+
\text{)}

+
\text{aff(}
+
\text{)}
+
\text{?}
\text{How doe we combine affinities }
\text{to get pixel-wise affinities for whole image}
Meeting with Florian
By Manan Lalit
Meeting with Florian
March 2
- 260

