
Alzheimer's disease project
ECSIT expression in
eukaryotic cells
Crystallography lab meeting, 6/9/16
presented by
S. Acajjaoui
Canonical isoform
ECSIT1 (50.2 kDa)
1 -

Sensing
No significant cell damage
Reversible cell damage
Irreversible cell damage
ECSIT protein in Alzheimer's disease
We are focusing on ECSIT when involved in mitochondrial pathway, and specifically for the Complex I assembly as chaperone

- cytoplasmic and nuclear signalling protein
- Nterminal sequence targeting the mitochondria
ECSIT: Evolutionarily Conserved Signalling Intermediate
in Toll-pathway
Our hypothesis:
We speculate that ECSIT, in collaboration with other Complex I assembly factors, are involved in Alzheimer’s disease pathogenesis, since defects in CI are observed in the early stages of the disease.
Our hypothesis:
ECSIT would function as an integrating signaling hub to maintain cell homeostasis in response to amyloid-beta or oxidative damage signals.
Failure to repair would generate severe mitochondrial damage and ultimately activate apoptotic mechanisms, promoting synaptic dysfunction and neuronal death.
Investigation of ECSIT function in eukaryotic cells

1. Defining a target
- ECSIT full length ? Part of full length ? Isoforms ?
- Try to find a potential partner : how to evaluate the complex interaction ? Can we purify it ?
- Impact of overexpressing one of the targets ?
2. Finding good cells for our experiment
- cells that are able to reproduce
the AD pathology - easy to handle

We decided to carry out functional assays on established human cell lines (HEK and H4), and test protein-protein interactions in yeast (using the two-hybrid system – Y2H)
Mammalian cells (MC) experiments
HEK293T cells
Human Embryonic Kidney transformed with a large T antigen to help expression
Cells are adherent on a monolayer and easy to manipulate for DNA transfection and expression of our target
Unlike bacteria, MC have a deadline for using it (around pass 30)
They grow at 37 deg. with 5% CO2 and they are very sensitive to contamination.
H4 cells
Adherent cells from human brain suffering from neuroglioma.
we have to lines : wild-type (H4wt) and overexpressing the amyloid precursor protein (APP) (H4app)
Cancer cells: metabolic pathways might be already altered as compared to non-proliferative cells
Those cells are robust but transfection is (really) hard
Having the two lines enables us to analyze the effect of amyloid formation in vivo, being one of the best cell models to study Alzheimer’s
MC experiments
Aim: obtain recombinant protein expressed in human cells
Study the possible protein maturation (ubiquitination...)
MC experiments: how to handle it?

- Cells are split and expanded as adherent cultures






2. we create our library
At low passage number, “ young” cells are frozen and kept in liquid nitrogen as stock
3. an aliquot is taken for starting study



When cells are too “old” : we throw them away and we take a new sample from the library.


After thawing cells, they need to be stabilized (3-5 passages) before use
Transfection
Transfection protocol

Test 3 reagents:
- PEI
- Lipofectamine3K
- XtremGene
Protein analysis
by Western blot
- PEI is the most suitable reagent for large-scale transfection
Results: Production of recombinant protein ECSIT
- After 24h-48h gene transfection, we harvest the cells are lyse them chemically. We extract the soluble content to analyze protein expression by WB or purify it by column chromatography

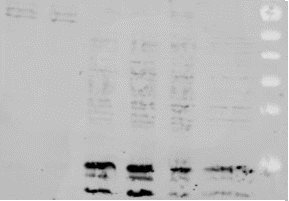

55 kDa
40 kDa
55 kDa
10 kDa
Ubiquitin lost during purification
Results : Ubiquitination site and exact mass
Mitochondrial signal need to be added : lysine 96

- H4 cells – DNA transfection issues

With standard protocol :
very low efficiency and cells die

H4 cells transfection improvement
- Working with 3:1 PEI:DNA (instead of 2:1 for HEK cells)
- Starving cells 24h before transfection: leave cells without FBS, Antibiotics before transfection
- Add concentrated DNA before transfection: remove all media from cells, drop complex
- DNA: PEI and leave it for 2h then add half media without FBS O/N
- the day after add half normal media (with 10% FBS)


H4 cells: still some work to do
Quantity of protein obtained is not enough
We need to work on purification protocol to avoid degradation of ECSIT


Protein degrades quickly...

Next step: working with this plasmid, ECSIT inserted between 2 tags could be protected from degradation
Yeast to Hybrid (Y2H)
Y2H: How does it work ?

If X and Y interact, a functional transcription factor is formed
We will work on histidine induction because it is the more sensitive.
We will use 3AT as an inhibitor of the activation to estimate the affinity.
Y2H: screening

We select yeast in agar plate lacking a specific amino acids: leucin and tryptophan, to select double transforming cells and histidine for the screen.

Agar plate w/o Leu&Trp
Y2H: hybrid assay
We tested 10 different constructs:

Y2H hybrid assay preliminary results
Results need to be confirmed by repeating screening, and if possible screen other plates (beta Gal assay, Uracil screen)


Thanks to Romain, Serena, Gabriele and Montse for your hard work.
Thanks for your attention !
Any questions ?
samira_labmeeting_6sep2016
By Matias Guijarro
samira_labmeeting_6sep2016
- 476



