Massively parallel implementation in Python of a pseudo-spectral DNS code for turbulent flows
Assoc. Professor Mikael Mortensen
Department of Mathematics, University of Oslo
Center for Biomedical Computing, Simula Research Laboratory
at EuroScipy 2015
Cambridge, UK, 28/8 - 2015
Direct Numerical Simulations of turbulence are performed with low-level Fortran or C
3D + time and HUGE resolution
Serious number crunching
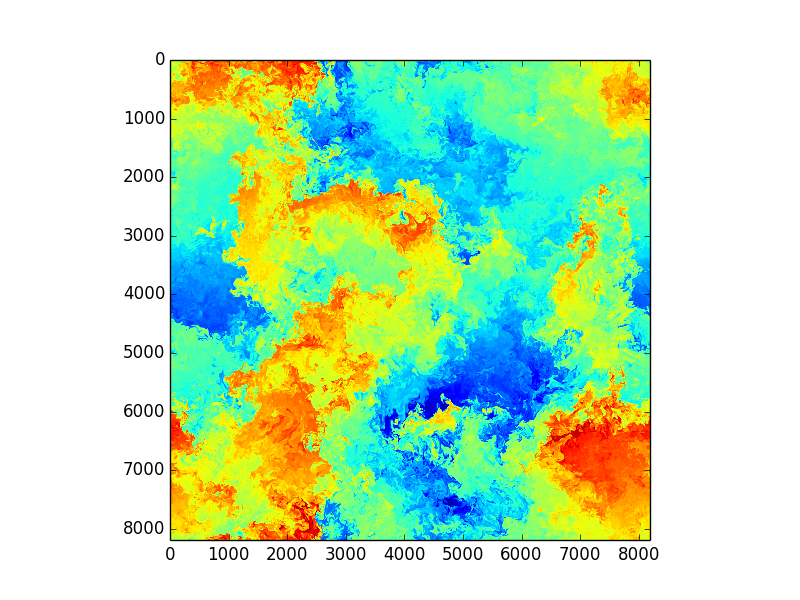
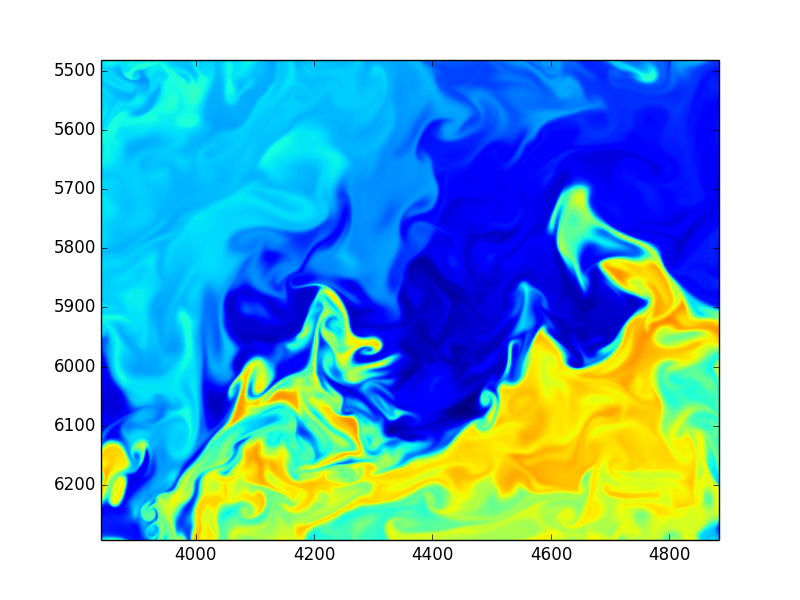

Images from huge DNS simulations by S. de Bruyn Kops, UmassAmherst, College of Engineering
Is it possible to write a competitive, state of the art turbulence solver in Python?
From scratch?
(Not just wrapping a Fortran solver)
Using only numpy/scipy/mpi4py?
Can this solver run on thousands of cores?
With billions of unknowns?
As fast as C?
Turns out 100 lines of Python code can get you almost there!

Turbulence is governed by the Navier Stokes equations
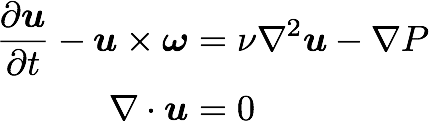

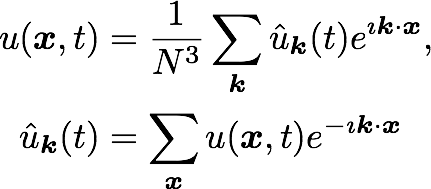
Triply periodic domain
Fourier space
Navier Stokes in spectral space
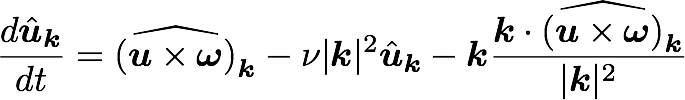
Eliminate pressure and solve ODEs:
Pseudo-spectral treatment of
Explicit timestepping (4th order Runge-Kutta)

Classical pseudo-spectral Navier-Stokes solver
The kind that has been used to break records since the day of the first supercomputer
Implementation
A few computational details
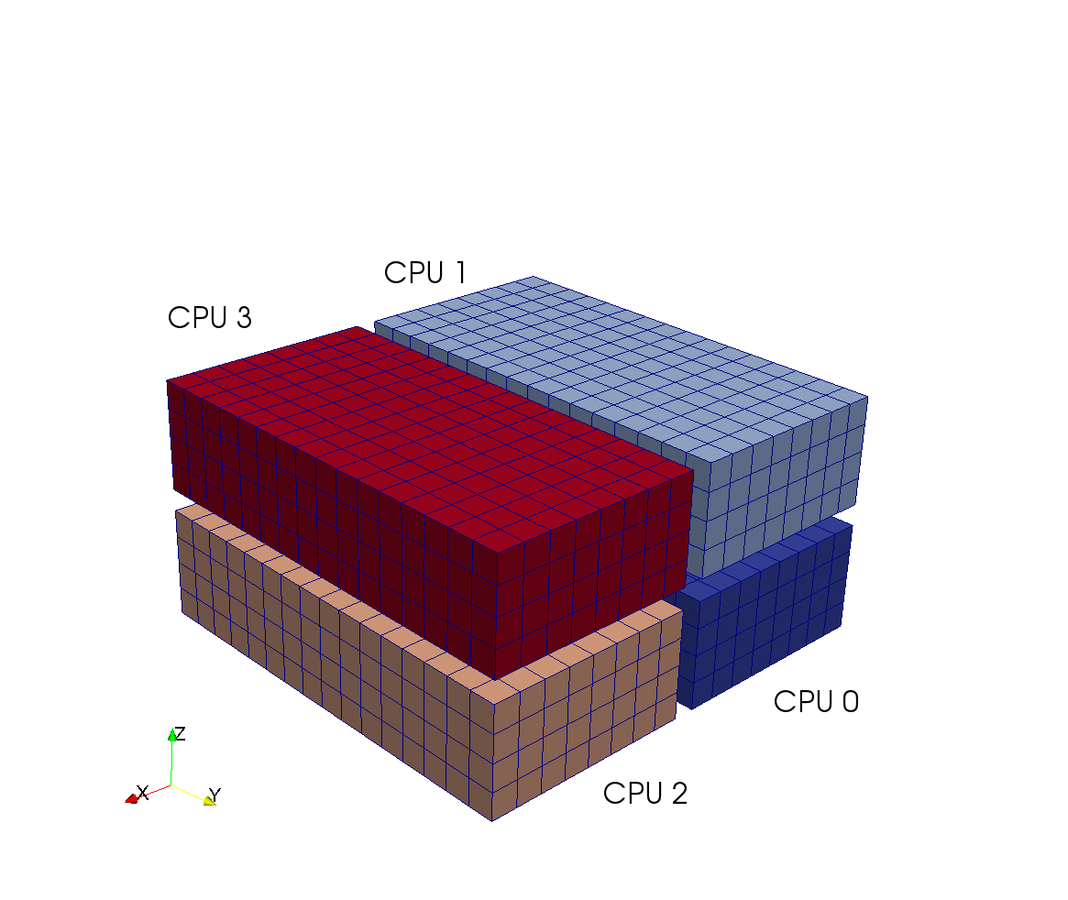
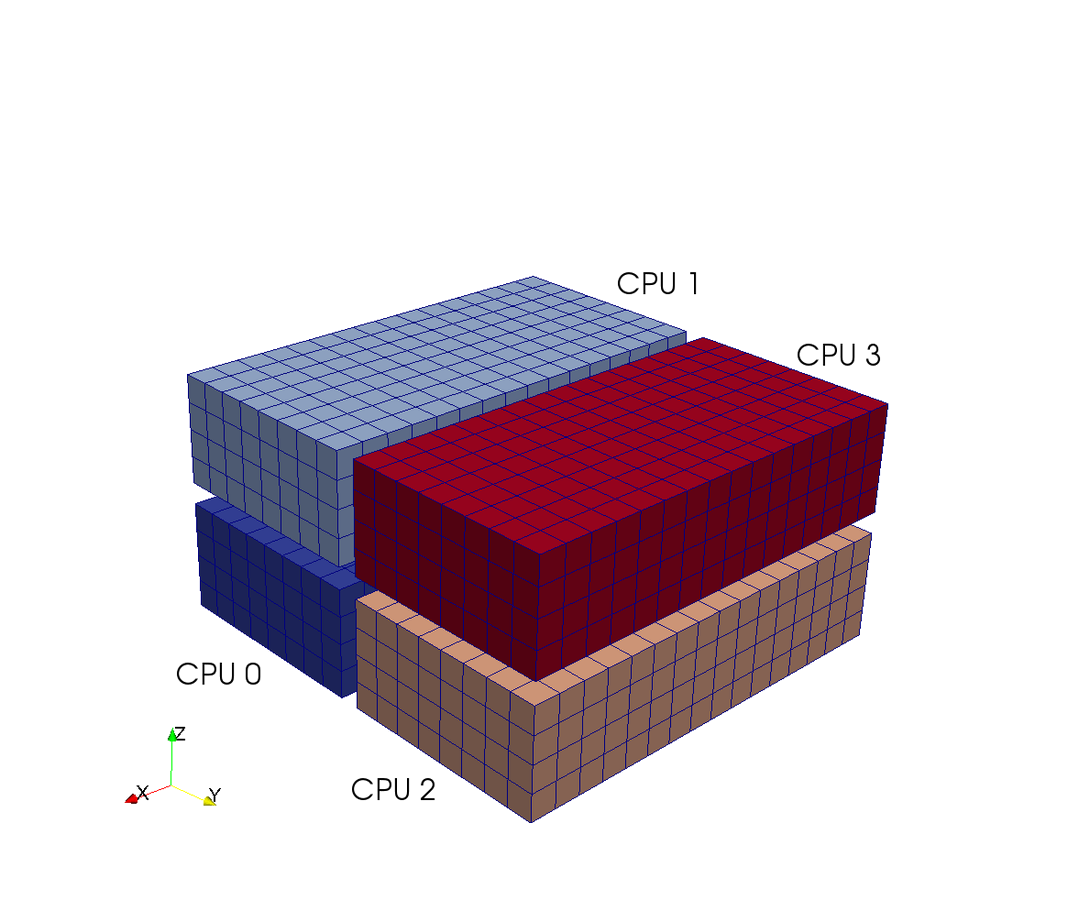
A computational box (mesh) needs to be distributed between a (possibly) large number of processors
Mesh size:
Pencil
Slab
CPUs
CPUs
mpi4py
wraps almost the entire MPI library and handles distribution of numpy arrays at the speed of C
from mpi4py import MPI
comm = MPI.MPI_COMM_WORLD
print "Hello World from rank", comm.Get_rank()>>> mpirun -np 1000 python HelloWorld.py
Hello World from rank 0
Hello World from rank 1
Hello World from rank 3
...MPI is basically needed to do 3D forward and backward FFT
HelloWorld.py:
3D FFT with MPI
Slab decomposition (max N CPUs)
def fftn_mpi(u, fu):
"""FFT in three directions using MPI."""
Uc_hatT[:] = rfft2(u, axes=(1,2))
for i in range(num_processes): # Np = N / num_processes
U_mpi[i] = Uc_hatT[:, i*Np:(i+1)*Np]
comm.Alltoall([U_mpi, mpitype], [fu, mpitype])
fu = fft(fu, axis=0)
return fuWith serial pyfftw (wrap of FFTW) and
5 lines of Python code!
real FFT in z, complex in y
No MPI required
Transpose and distribute
Then FFT in last direction
There are a few low level 3D FFT modules with MPI (e.g., libfftw, P3DFFT) but none that have been wrapped or is easily called from python
Need to adapt single core FFT
(numpy.fft, scipy.fftpack, pyfftw)
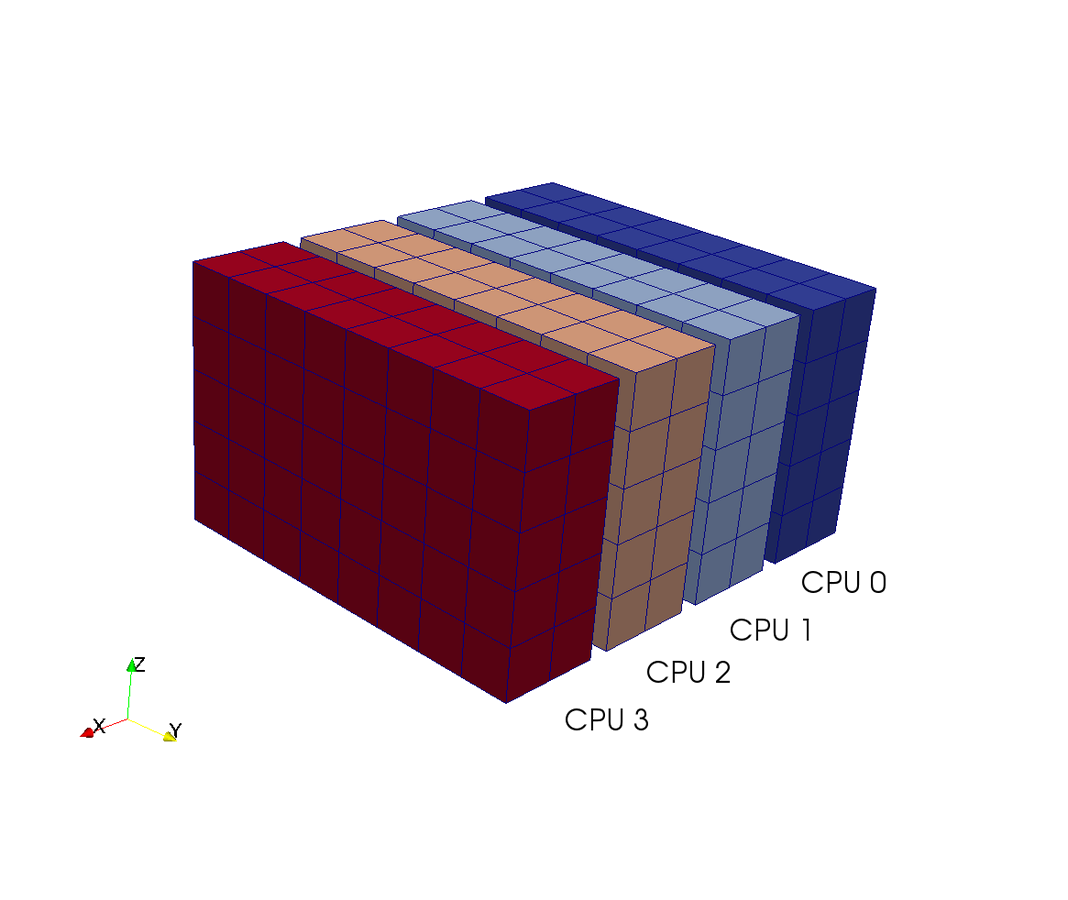
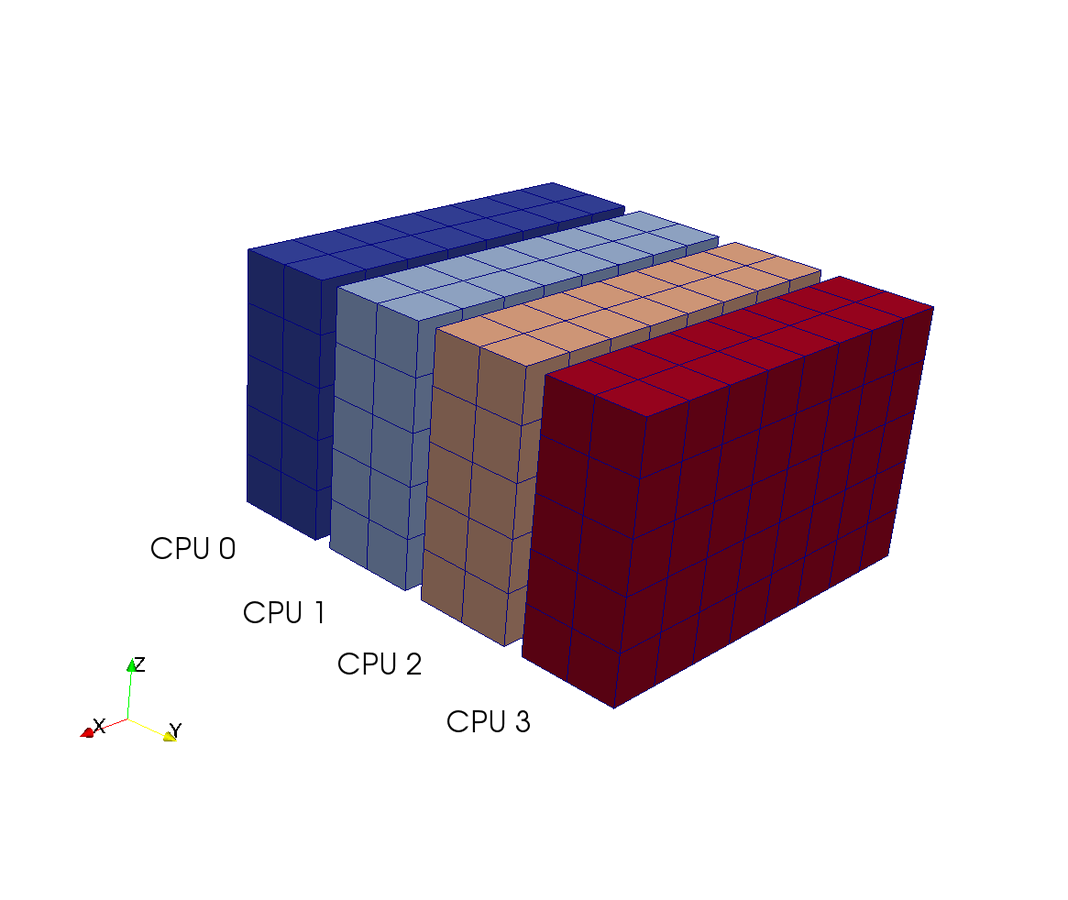
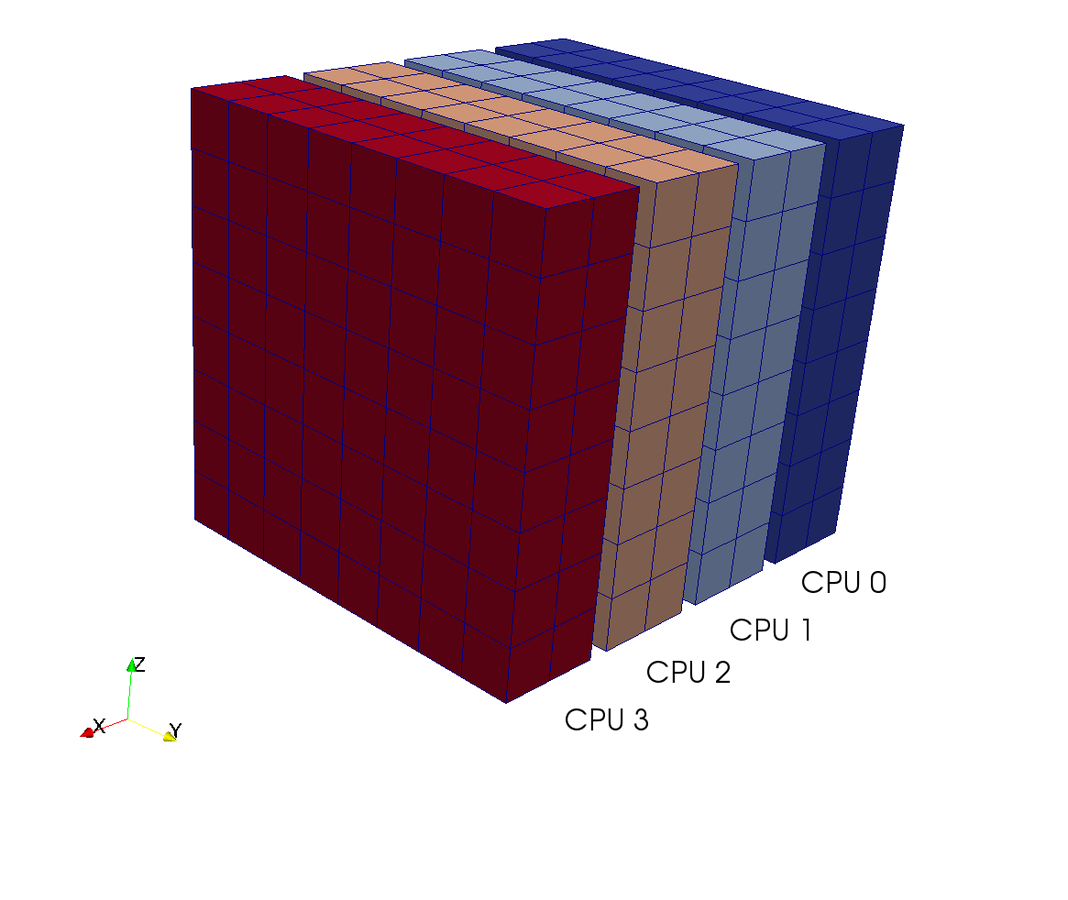
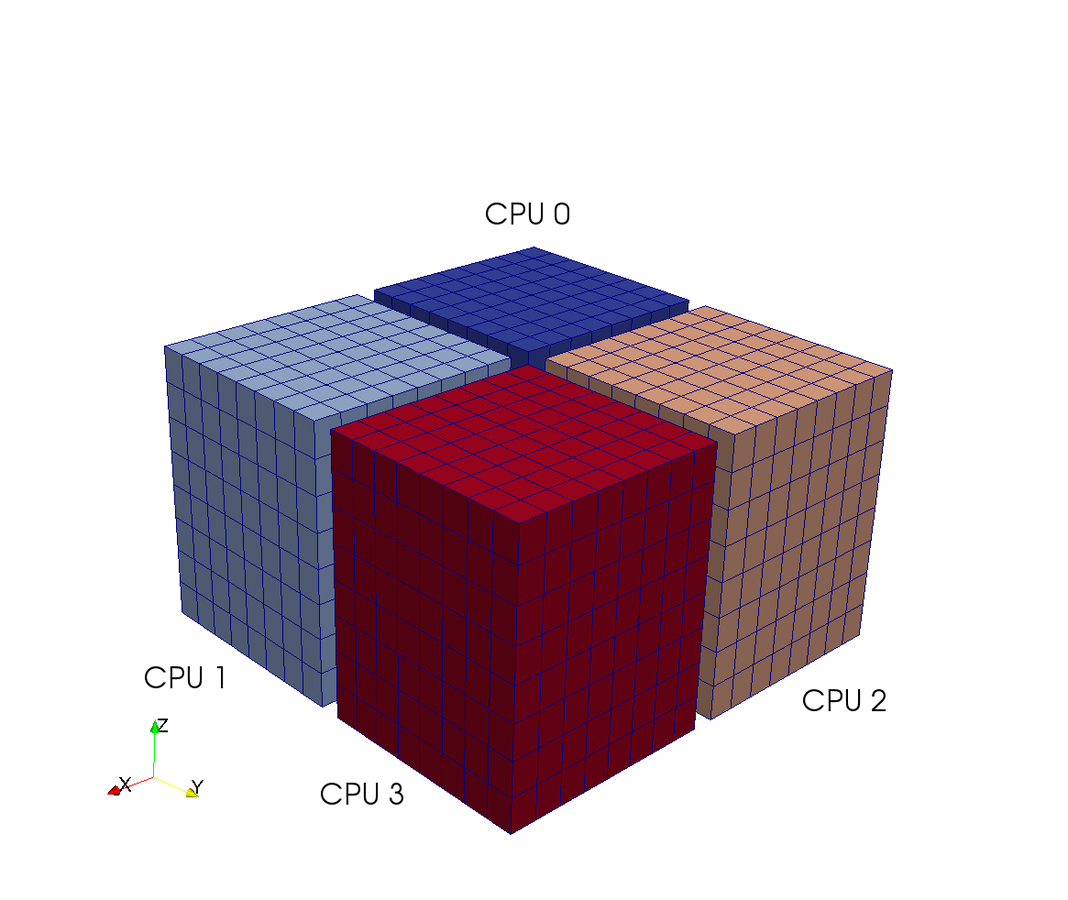
Pencil decomposition
Up to CPUs
10 lines of Python code
to do 3D FFT with MPI
rfft(u, axis=2)
Transform
fft(u, axis=1)
Transform
fft(u, axis=0)

Numpy ufuncs responsible for most remaining cost
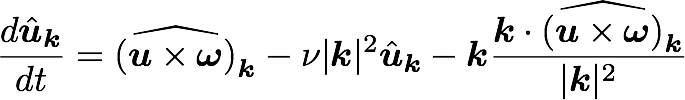
# Preallocated distributed arrays. Create array of wavenumbers
K = array(meshgrid(kx, ky, kz, indexing='ij'), dtype=float)
K2 = sum(K*K, 0, dtype=float) # Wavenumber magnitude
dU = zeros((3, N[0], Np[1], Nf), dtype=complex) # Rhs array
# Subtract contribution from diffusion to rhs array
dU -= nu*K2*U_hatBesides FFT the major computational cost is computing the right hand side using element-wise numpy ufuncs (multiplications, subtractions over the entire mesh)
Very compact code
Explicit solver
Parallell scaling
Shaheen Blue Gene P supercomputer at KAUST Supercomputing Laboratory
Testing with a transient Taylor Green flow
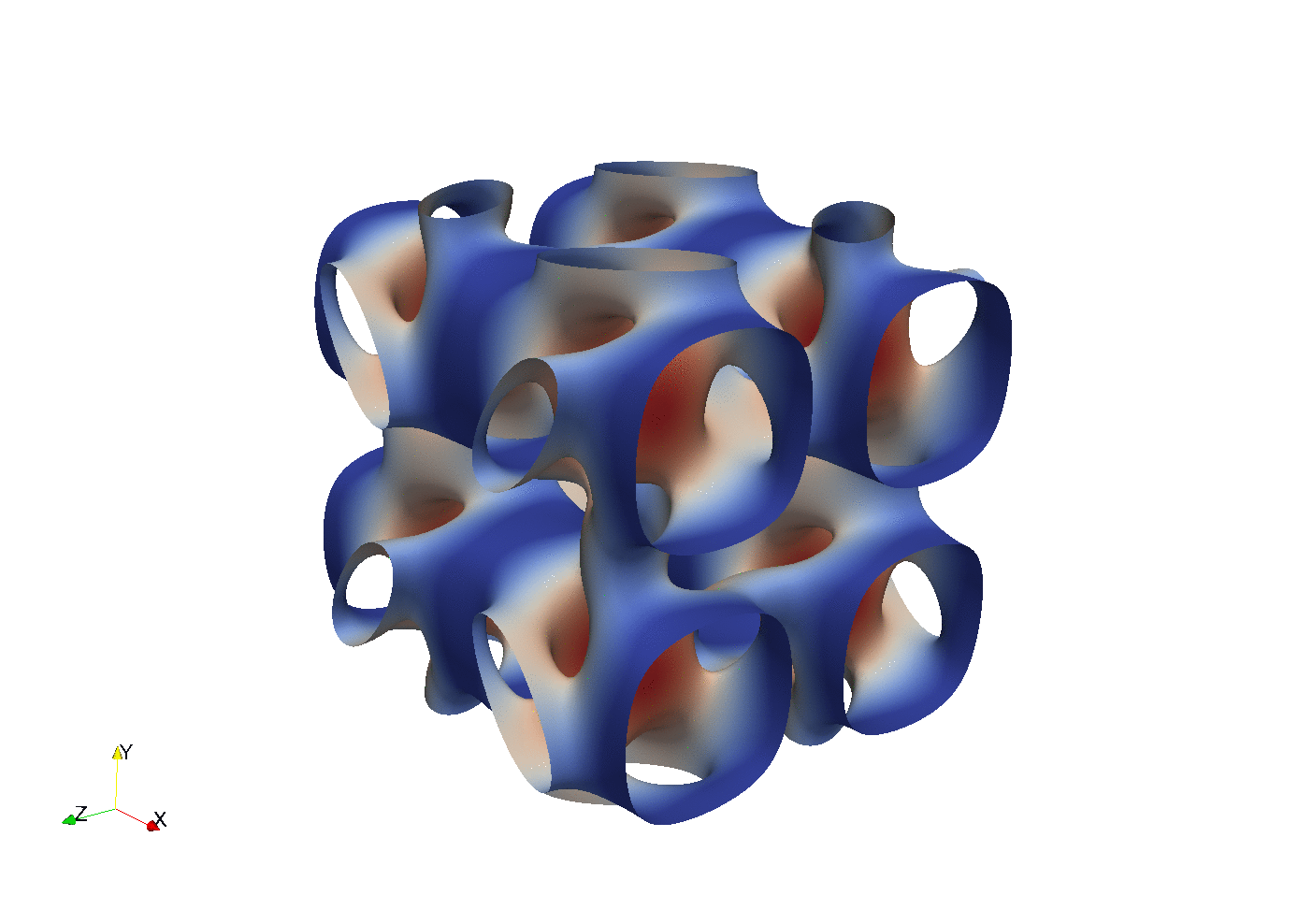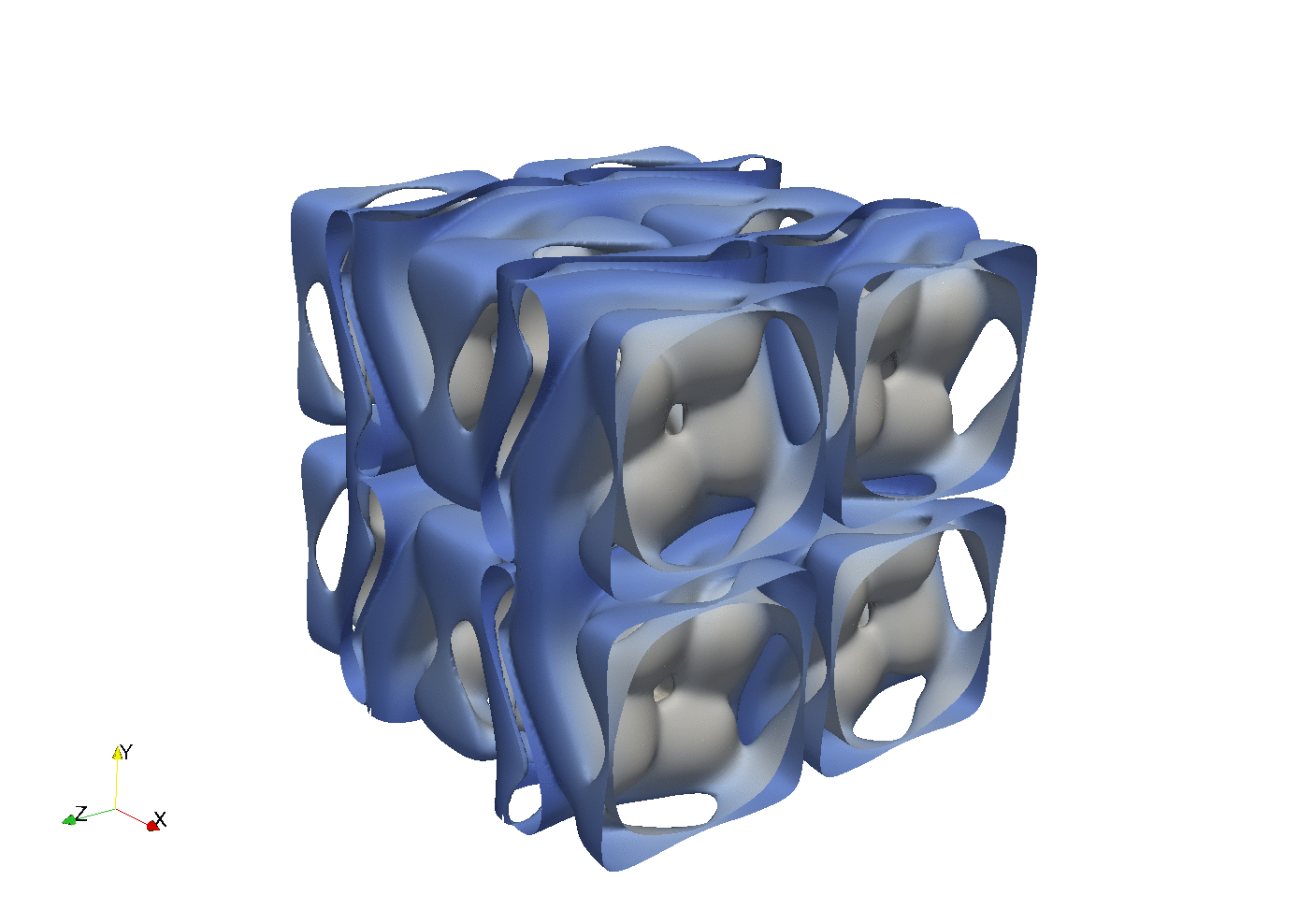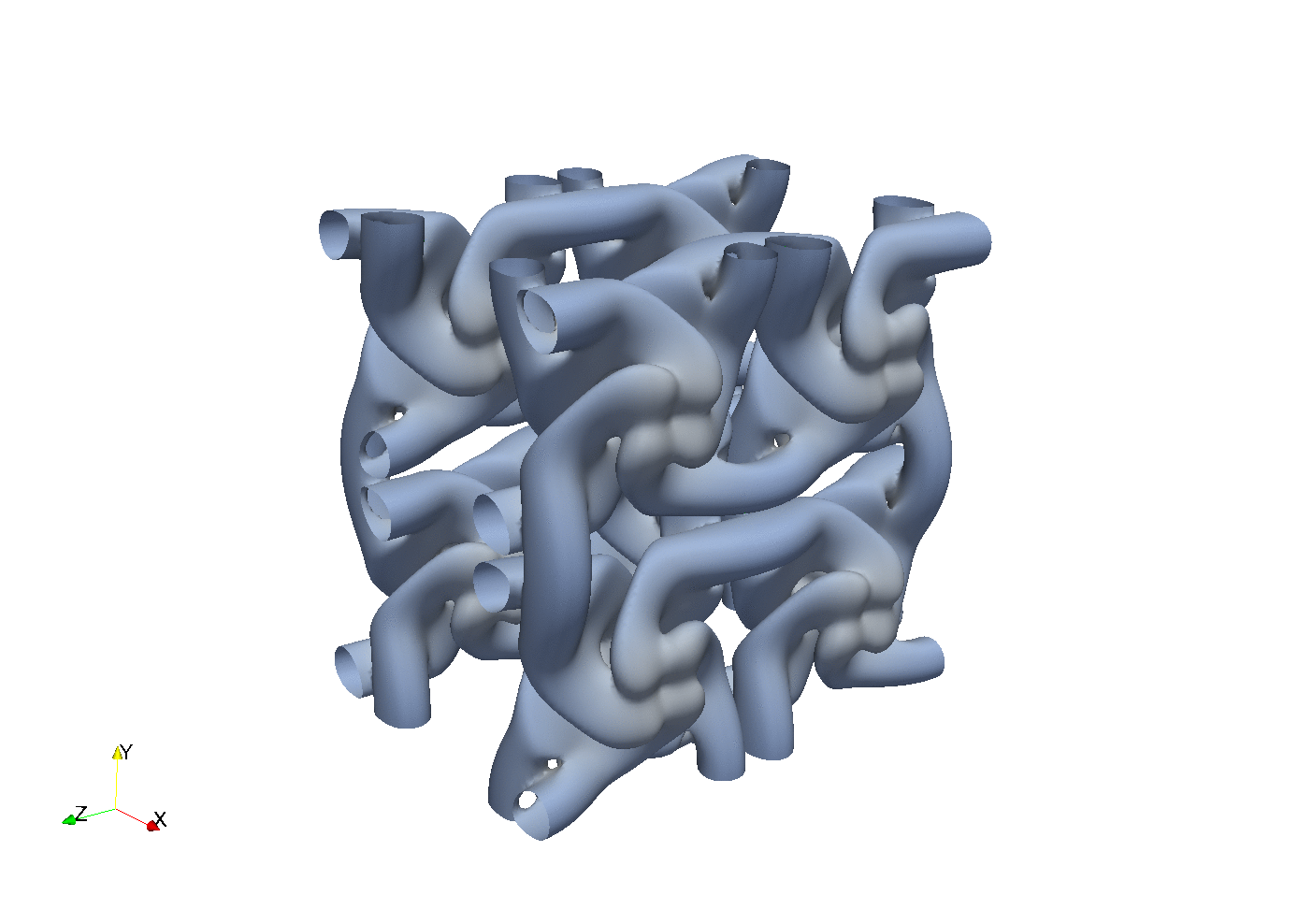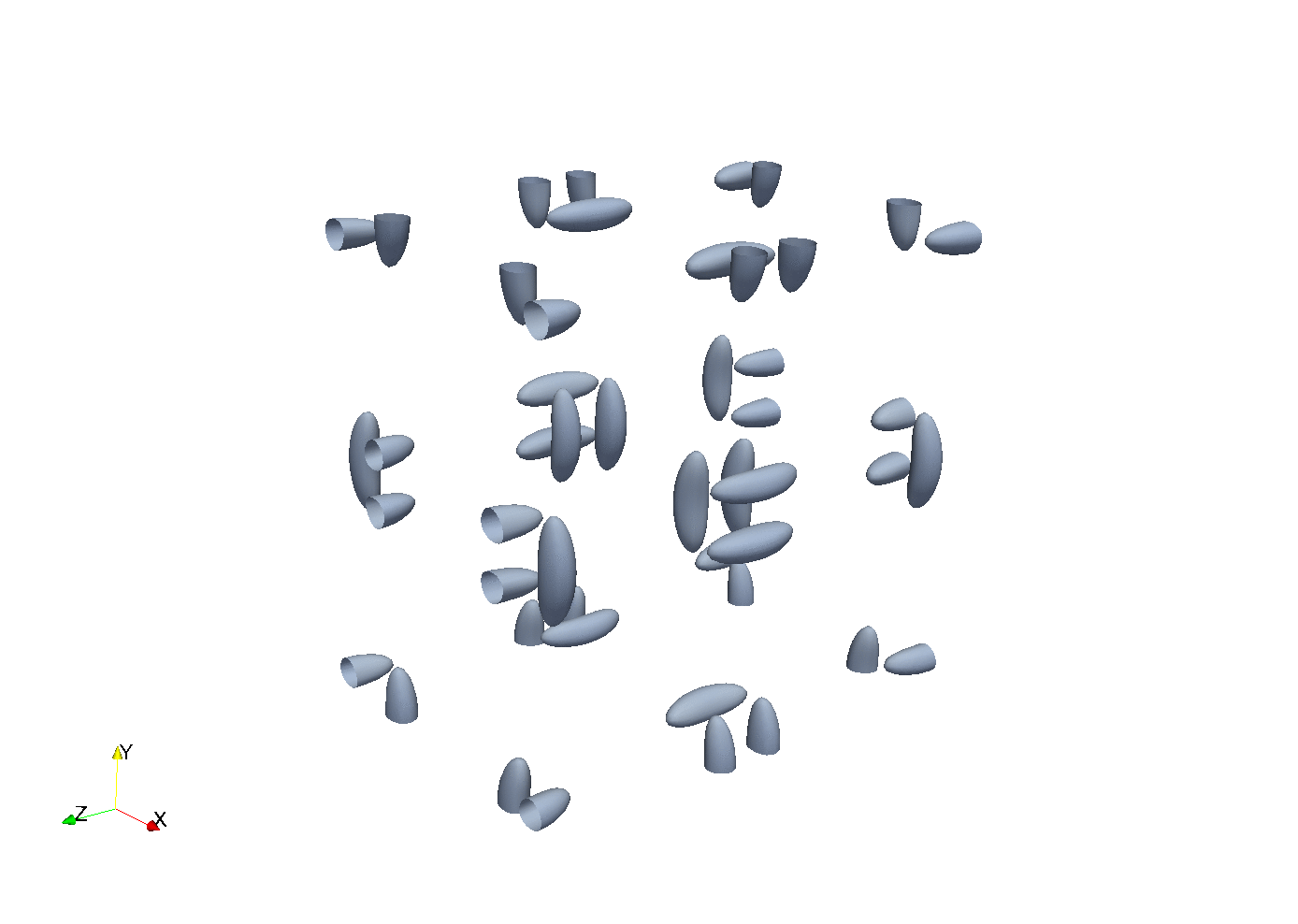
Enstrophy colored by pressure
The solver has been verified to be implemented correctly, but how does the pure numpy/mpi4py/pyfftw perform?
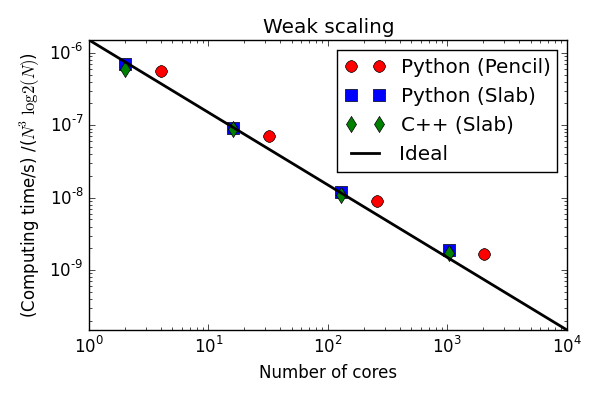
Parallell scaling
Shaheen Blue Gene P supercomputer at KAUST Supercomputing Laboratory
Testing the 100 lines of pure Python code
C++(slab) 30-40 % faster than Python (slab)
Python solvers struggling with the strong scaling
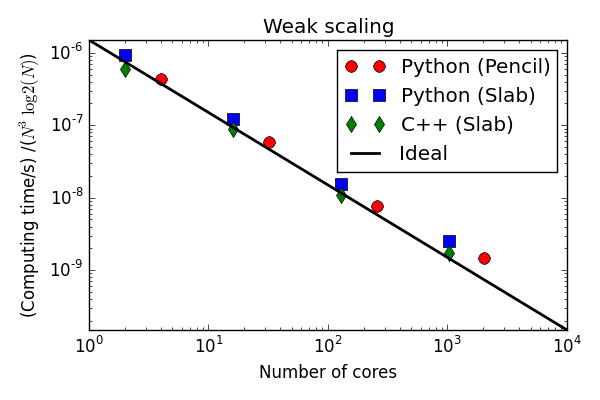
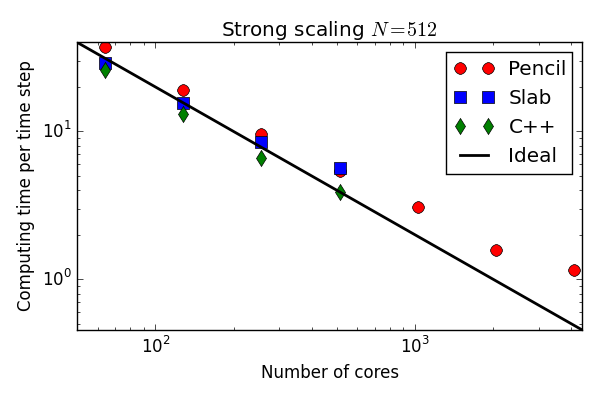
Why is the Python solver not scaling better?
def fftn_mpi(u, fu):
"""FFT in three directions using MPI."""
Uc_hatT[:] = rfft2(u, axes=(1,2))
for i in range(num_processes): # Np = N / num_processes
U_mpi[i] = Uc_hatT[:, i*Np:(i+1)*Np]
comm.Alltoall([U_mpi, mpitype], [fu, mpitype])
fu = fft(fu, axis=0)
return fuAll MPI communication is found in the 3D FFTs
There's a for-loop used to transform datastructures
that I could not get rid of...
Increasingly worse for larger number of processors
Why slower than C++?
The for-loop int the FFT is one obvious thing
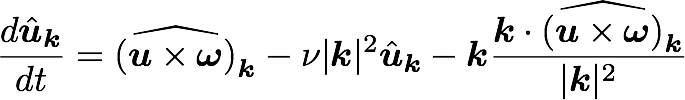
However, the ufuncs are the main cause,
at least for small number of CPUs
Numpy ufuncs not always great
def cross(a, b, c):
"""Regular c = a x b, with vector component in axis 0."""
#c = numpy.cross(a, b, axis=0) # Alternative
c[0] = a[1]*b[2]-a[2]*b[1]
c[1] = a[2]*b[0]-a[0]*b[2]
c[2] = a[0]*b[1]-a[1]*b[0]
return c
N = 200
U = zeros((3, N, N, N))
W = zeros((3, N, N, N))
F = zeros((3, N, N, N))
F = cross(U, W, F)
Consider the cross product (required for rhs):
Too many ufuncs (multiplications, subtractions) are being called and too many temp arrays are created
Commented out regular numpy.cross is also using ufuncs and is about the same speed
Getting rid of some temp arrays helps
def cross(a, b, c):
"""Regular c = a x b, with vector component in axis 0."""
c[0] = a[1]*b[2]
c[0] -= a[2]*b[1]
c[1] = a[2]*b[0]
c[1] -= a[0]*b[2]
c[2] = a[0]*b[1]
c[2] -= a[1]*b[0]
return c
Approximately 30% faster (for N=200),
but still too many ufuncs being called. Too many loops over the large 4D arrays a, b, c
Only viable solution
is to hardcode the cross product into one single loop
@jit(float[:,:,:,:](float[:,:,:,:], float[:,:,:,:], float[:,:,:,:]), nopython=True)
def cross(a, b, c):
for i in xrange(a.shape[1]):
for j in xrange(a.shape[2]):
for k in xrange(a.shape[3]):
a0 = a[0,i,j,k]
a1 = a[1,i,j,k]
a2 = a[2,i,j,k]
b0 = b[0,i,j,k]
b1 = b[1,i,j,k]
b2 = b[2,i,j,k]
c[0,i,j,k] = a1*b2 - a2*b1
c[1,i,j,k] = a2*b0 - a0*b2
c[2,i,j,k] = a0*b1 - a1*b0
return cCython, Numba or Weave will all do the trick
Numba and Cython perform approximately the same
and as fast as C++!
Approximately 5 times faster than original
Solver optimized with Cython
More than matches C++ on the Blue Gene P!
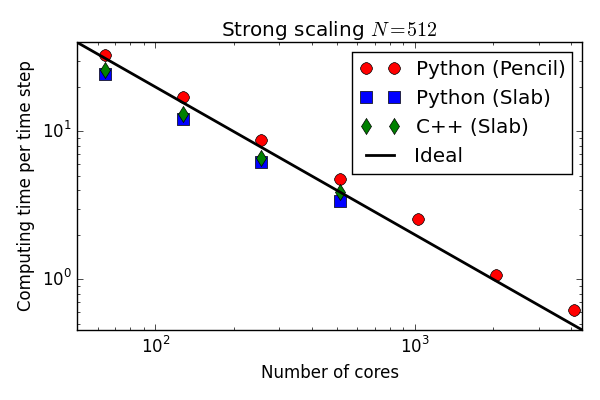
(Numba you say? You try to install Numba on a Blue Gene)
Final word of warning
Dynamic loading of Python can be very slow on supercomputers
- With regular Python 2.7 it takes 15 minutes just to get started on a full rack (4096 CPUs) on the Blue Gene/P
- In this work I have used Scalable Python (Python 2.6.7 with MPI-modified I/O) developed by J. Enkovaara and co-workers, which takes approximately 30 seconds to load.
Scalable Python Used to be at https://gitorious.org/scalable-python, but now I don't know?
Summary
- A pure Python DNS solver for turbulent flows can be created from scratch with less than 100 lines of Python code!
- This code can run on thousands of processors with billions of unknowns and almost (30-40% slower) as fast as pure C++
- With a few routines moved to Cython, the Python based solver matches the performance of C++
- Interested in helping out? Think you can do better?
- https://github.com/mikaem/spectralDNS
- Currently DNS solvers for incompressible/variable density Navier Stokes and Magneto Hydro Dynamics Isotropic (triply periodic) and channel flows
Thanks also to Hans Petter Langtangen at UiO/Simula
and David Ketcheson at KAUST
Thank you for your attention!
Massively parallel
By Mikael Mortensen
Massively parallel
- 1,919



