Structured Population Models
https://slides.com/nicolamueller/20190218ttbstrutured/edit



Bayesian Analysis requires to put priors on everything, which includes the tree
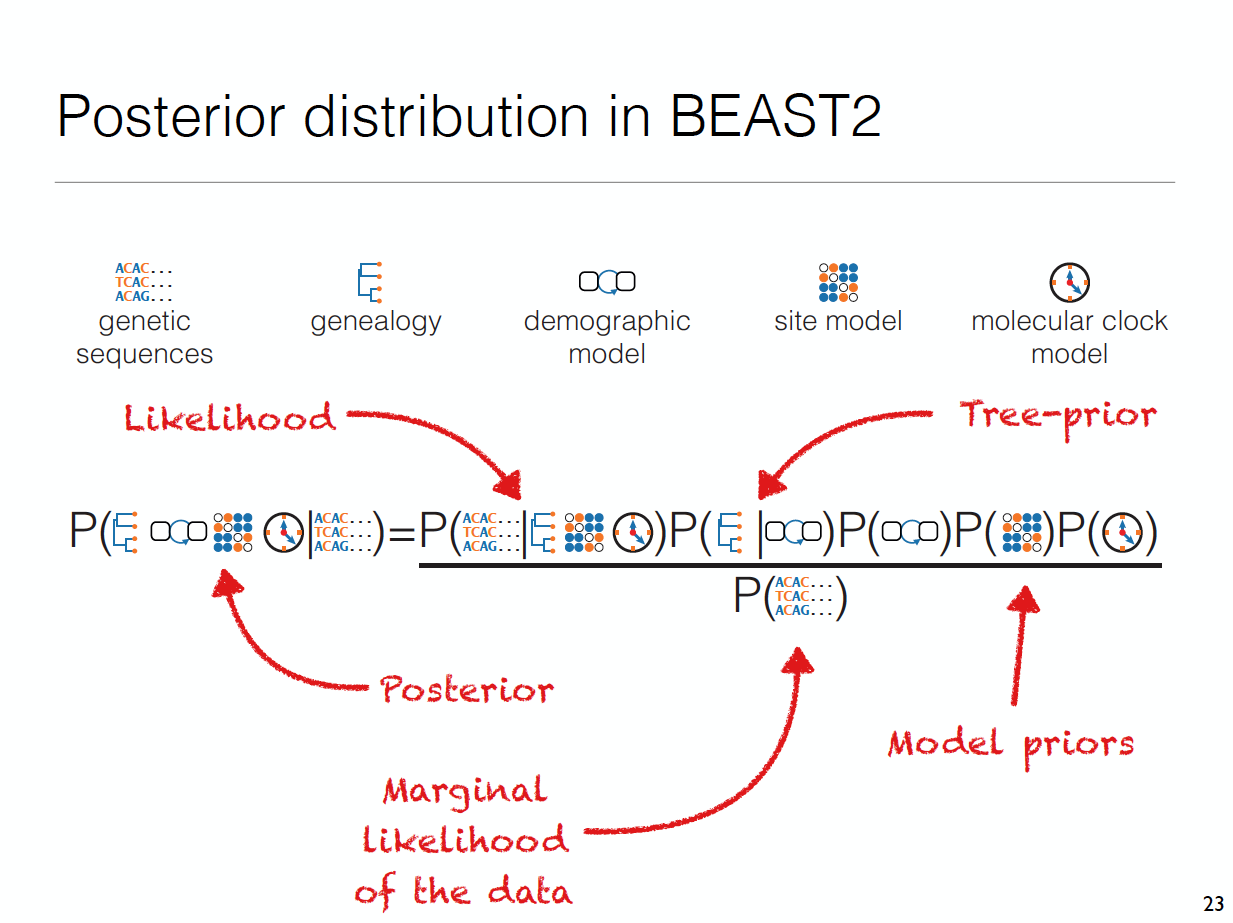
Tree generating models typically assume that populations are well mixed
This means that every lineage in a tree is equally likely to share a common ancestor with any other lineage.
Population structure, such as caused by geography, can violate that assumption
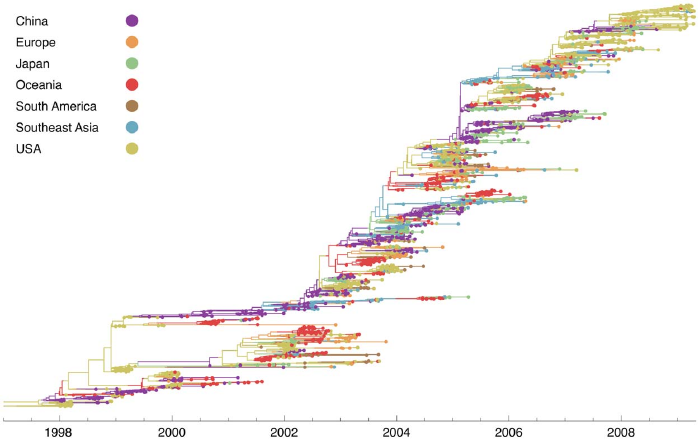
Bedford et al. (2015)
We have different approaches to deal with population structure
- Structured coalescent.
- Multi-Type birth-death models.
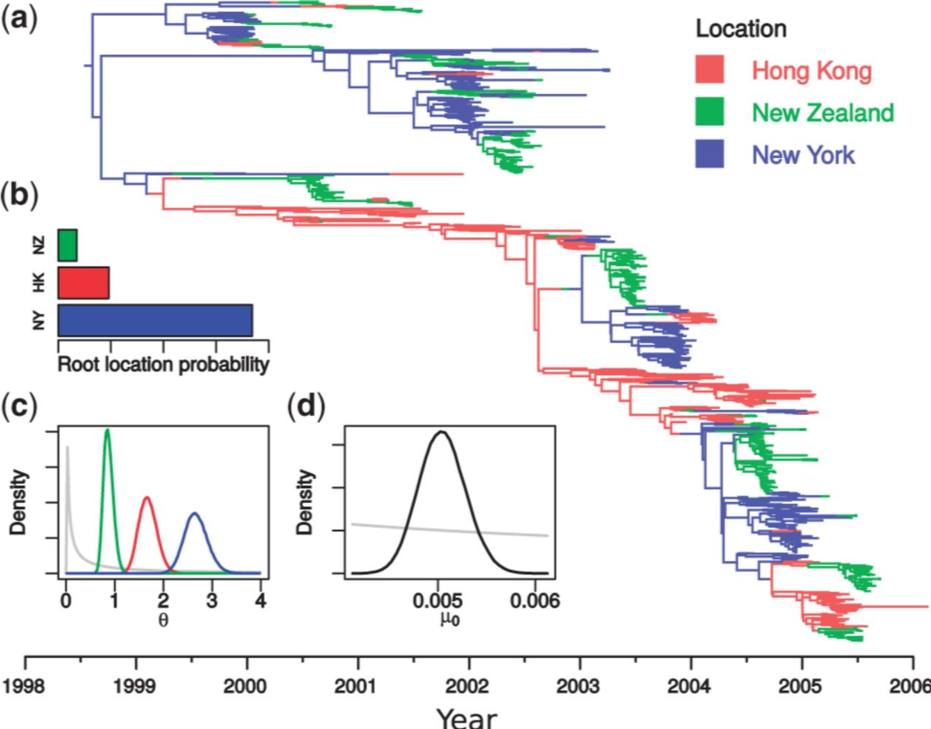
The structured coalescent models how lineages coalesce within and migrate between sub-populations

http://popgen.sc.fsu.edu/Migrate/Blog/Entries/2013/3/22_forward-backward_migration_rates.html
Spread of Influenza across different geographic locations inferred by MTT
Vaughan et al. (2014)
Population structure can have different origins
Dudas et al. (2018)
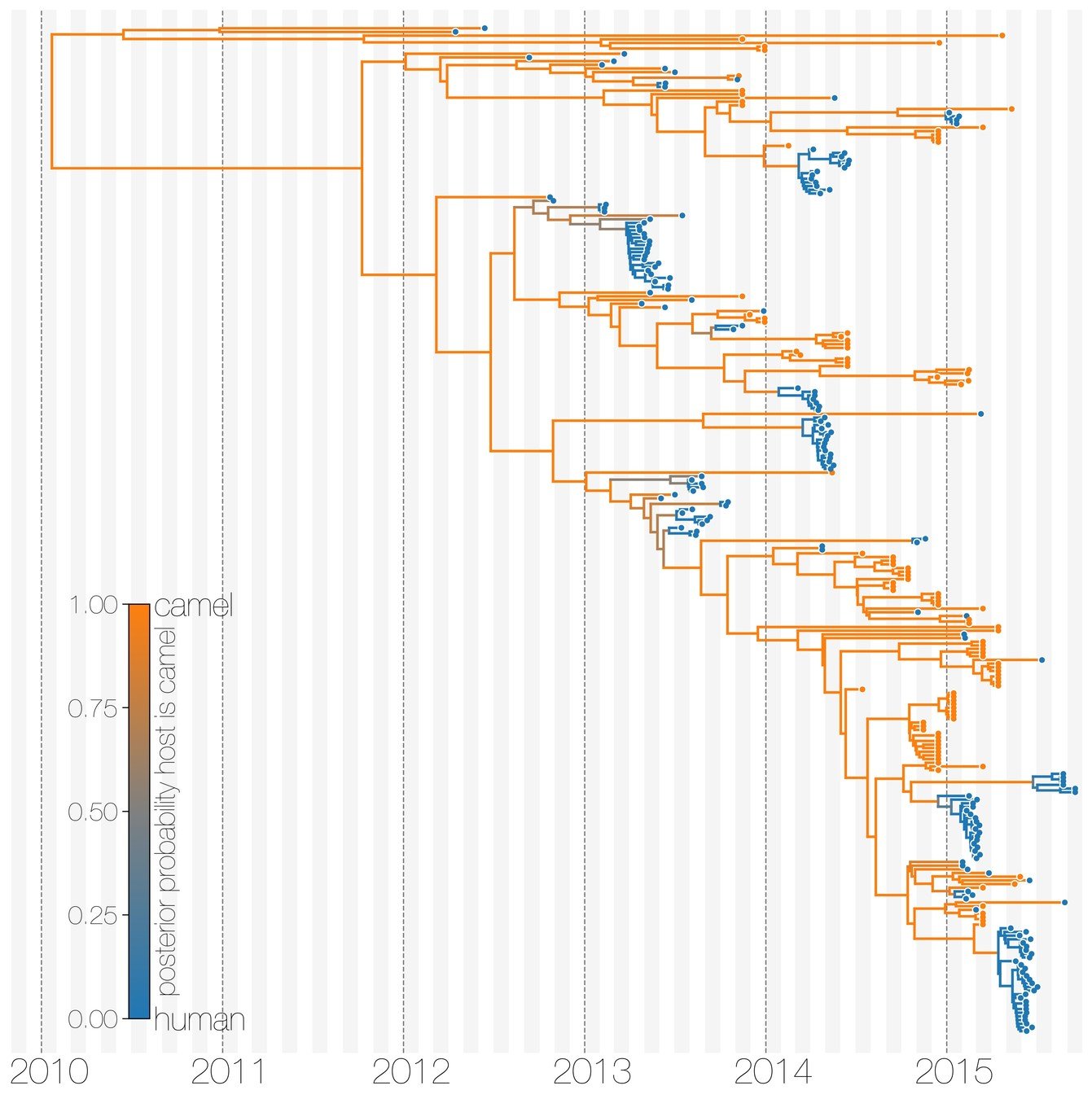
Viruses are considered to be part of different sub-populations depending on if they were isolated in humans or camels.
Exact structured coalescent are computationally limited to essentially 3 or 4 sub-populations
- This restriction originates from having to infer the colouring of phylogenetic trees (where lineages are over time).
- Integrating over every possible colouring allows to consider more complex scenarios.
In order to consider scenarios with more than a few sub-populations, we need approximate models
Different approximations perform differently when sampling is biased
Müller et al. (2017)

Different population sizes across different sub-populations can lead to wrongly inferred migration rates
Müller et al. (2017)
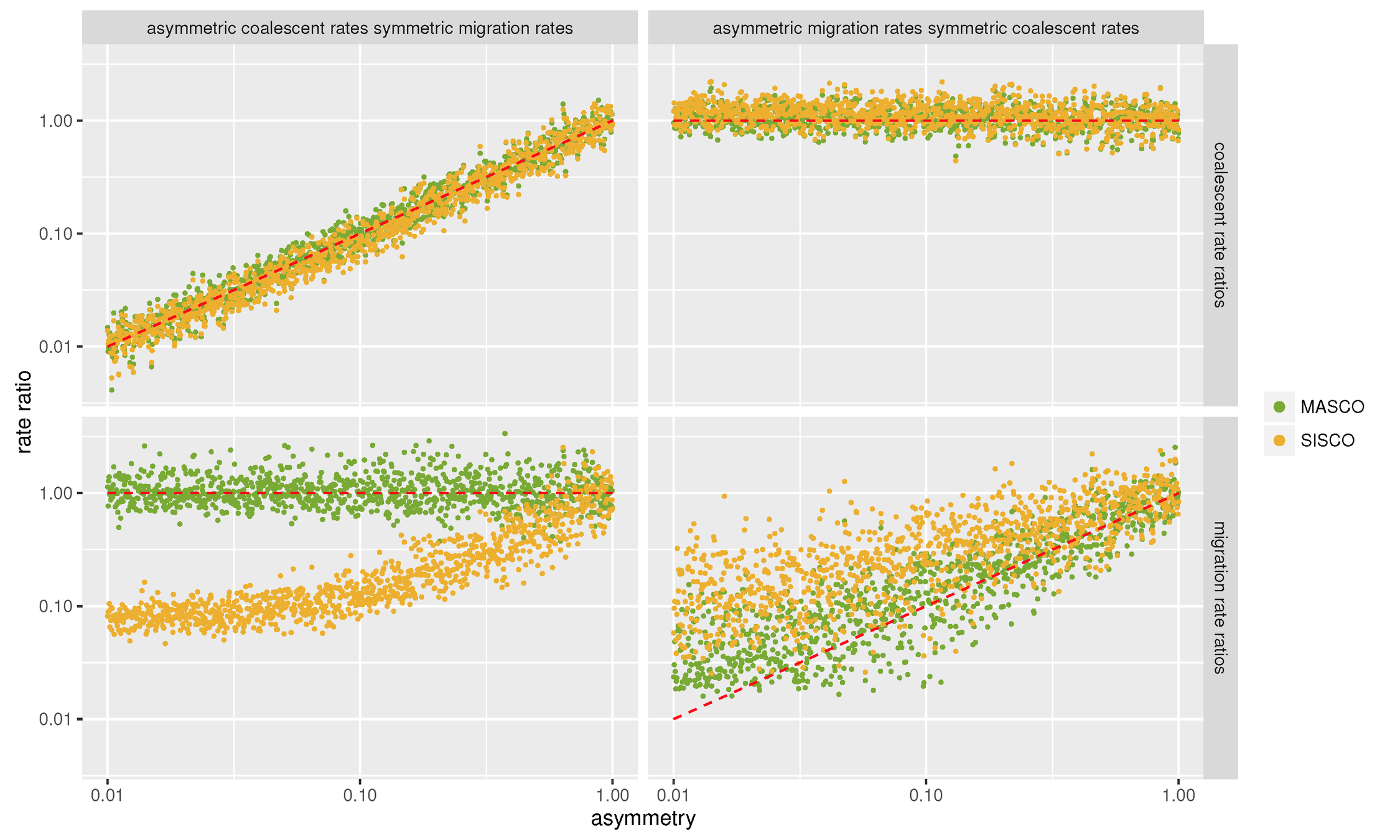
Mascot allows to analyse datasets with many different sub-populations
Müller et al. (2018a)

Effective population sizes and migration rates can be poorly informed or not of interest
- Instead of directly inferring these parameters, we say they are some combination of predictor data.
- Examples for predictor include geographic distance between sub-populations or weekly case data.
- MASCOT-GLM allows to infer which predictors and to what degree they inform effective population sizes and migration rates.
Müller et al. (2018b)
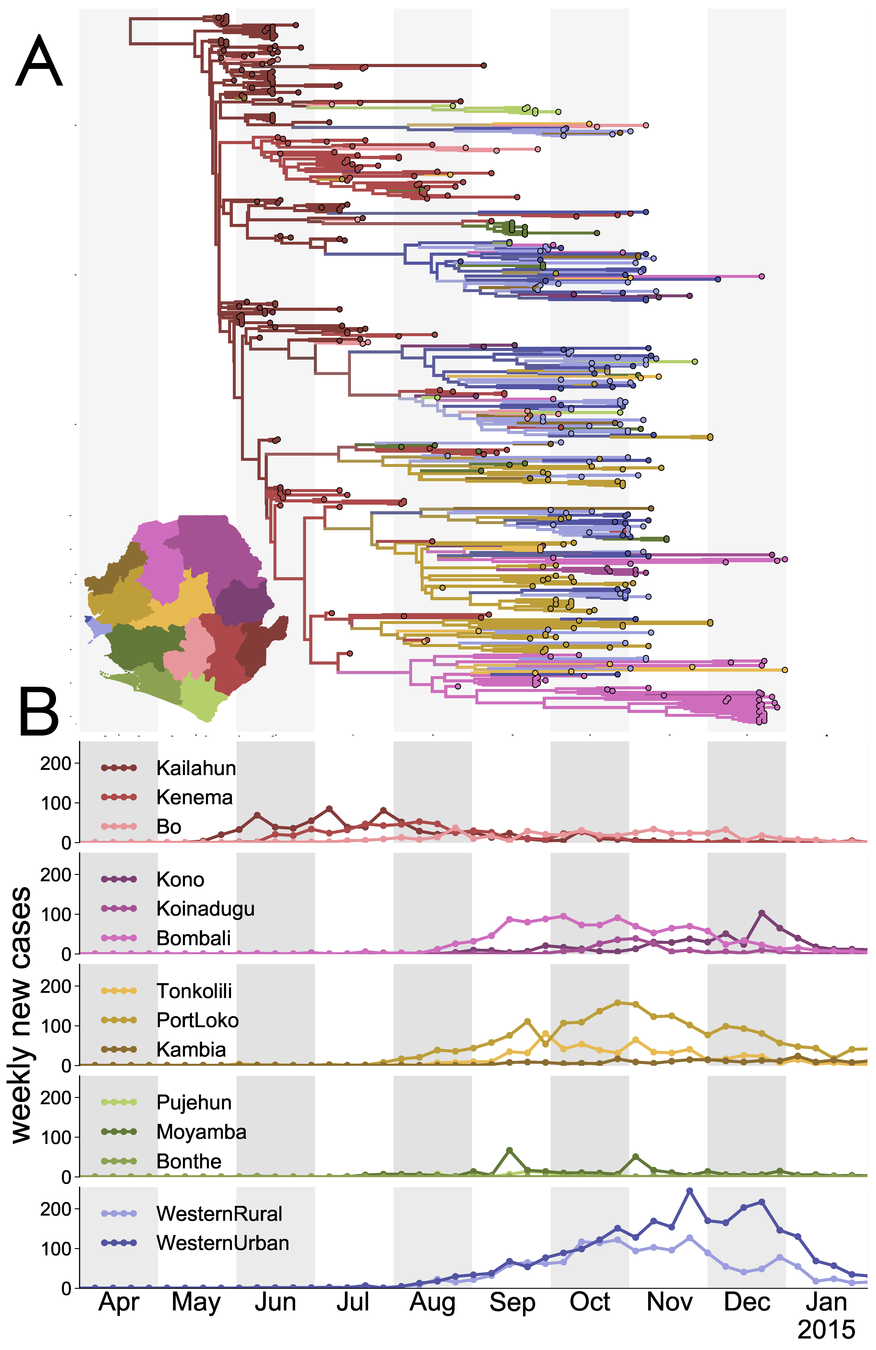
MASCOT allows to jointly reconstruct migration and coalescence patterns of EBOV from time varying predictor and genetic sequence data.
Müller et al. (2018b)
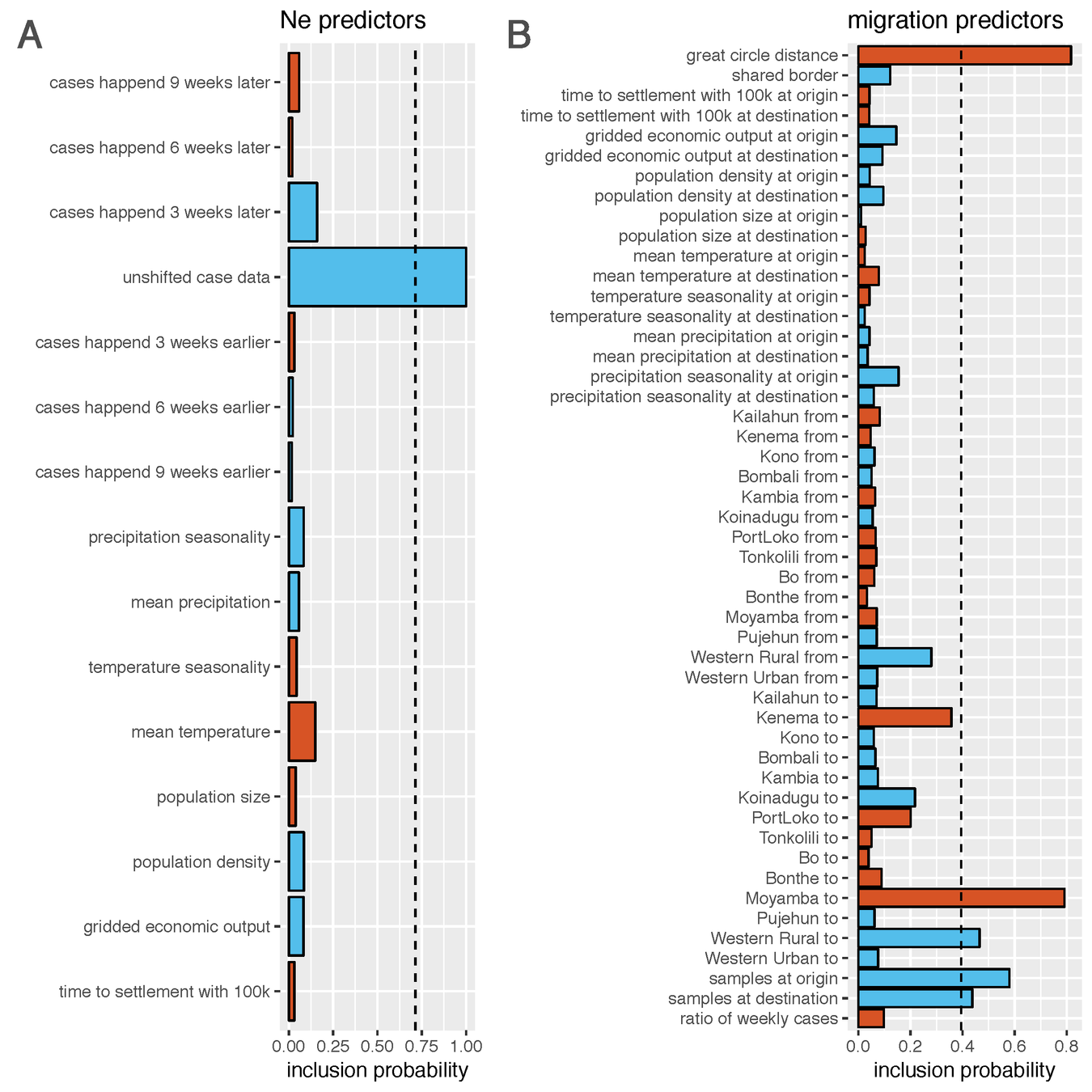
- Weekly case data is inferred to be the best predictor of effective population sizes through time and space
- Distances between different locations are inferred to be the best predictor of migration rates
Multi-Type Birth death models model the birth, death and sampling of lineages within and the migration of lineages between sub-populations
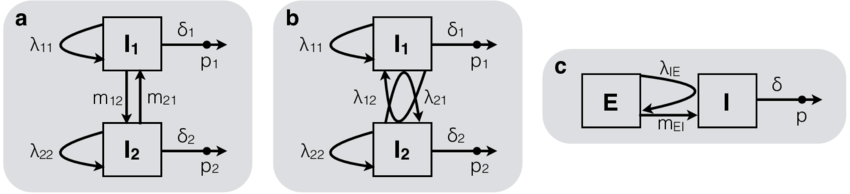
Kühnert et al. (2016)
Multi-Type Birth-Death models can be used to study migration patterns
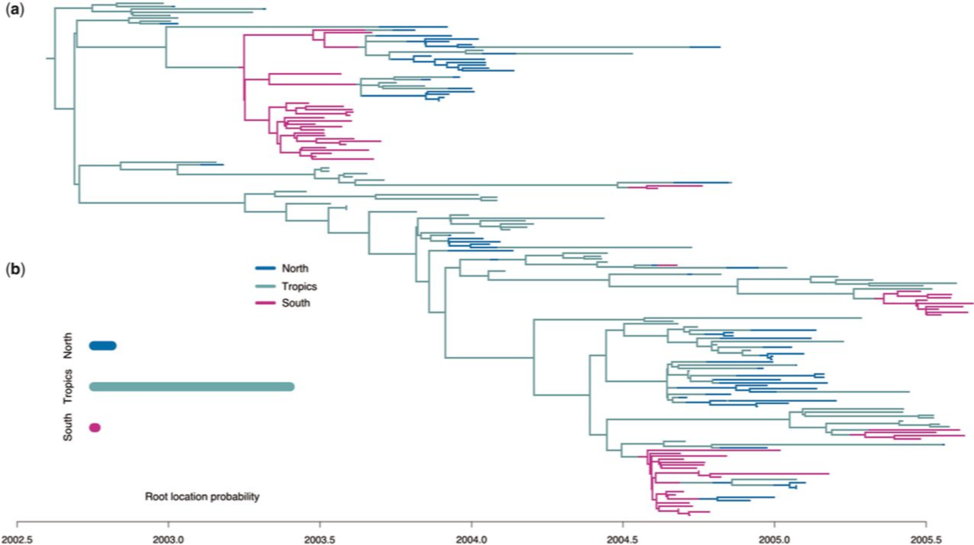
Kühnert et al. (2016)
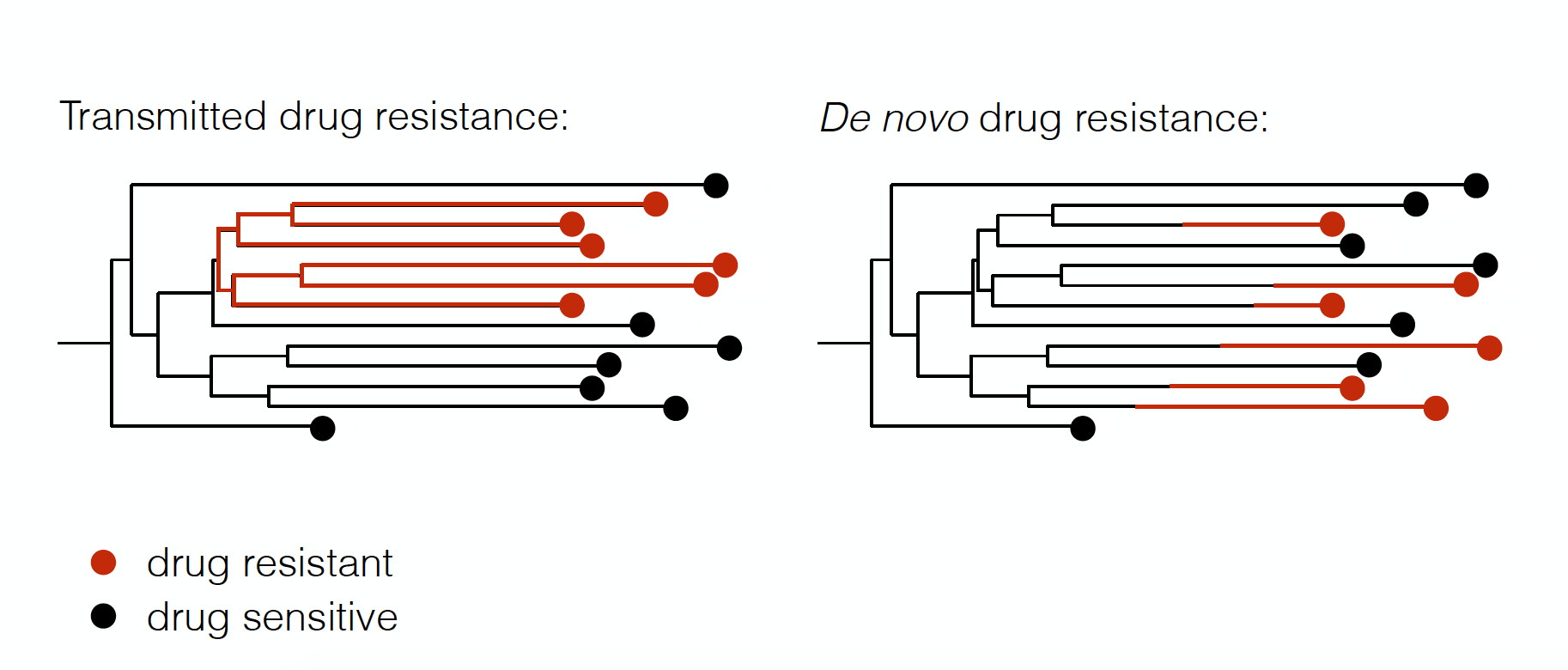
The same models can be used to study the fitness effects of drug resistance mutations
Kühnert et al. (2018)
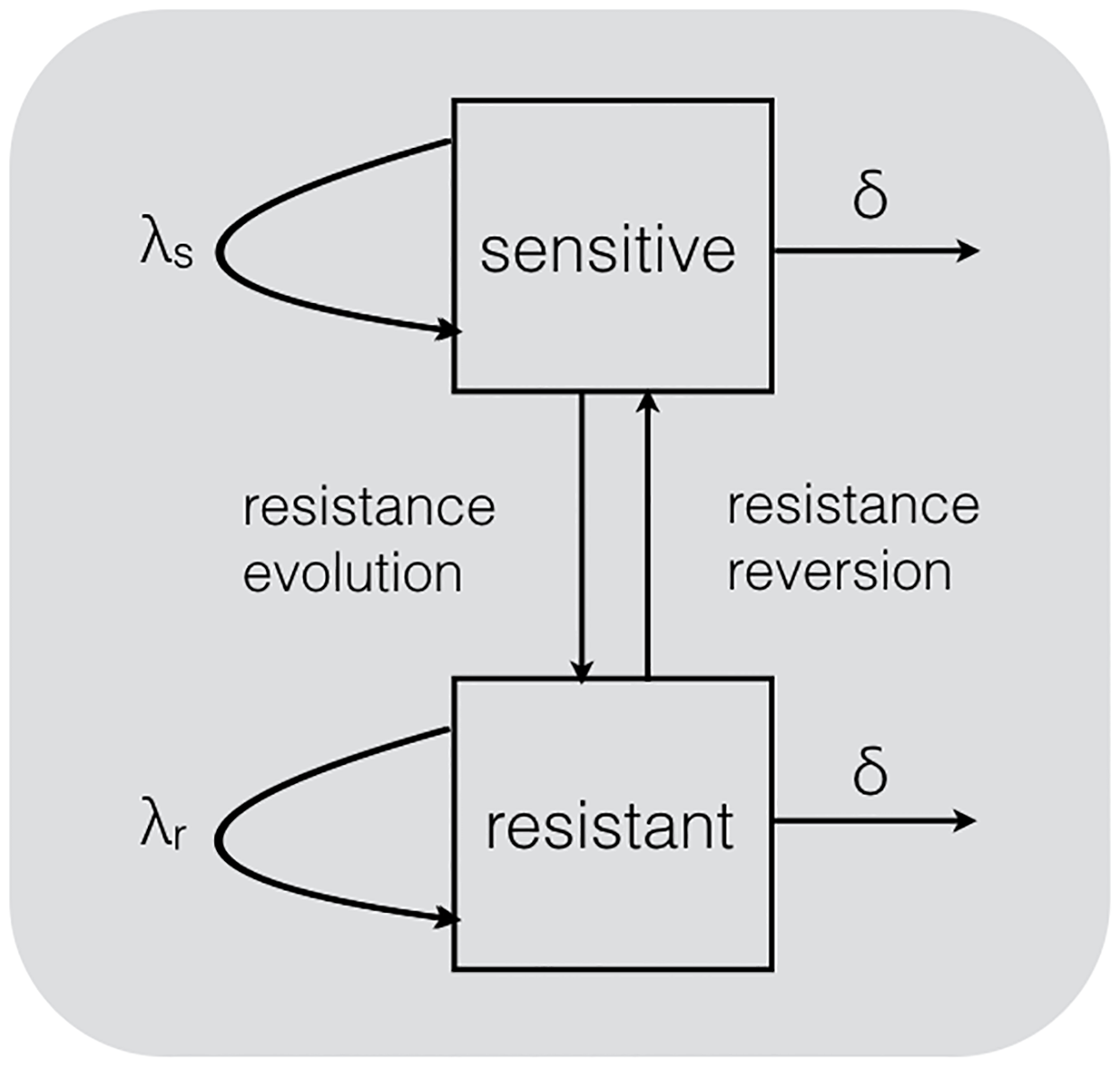
The same models can be used to study the fitness effects of drug resistance mutations
Questions?
Some References
- Theory behind Mascot and relationship between structured coalescent approximations: https://doi.org/10.1093/molbev/msx186
- Mascot-GLM: https://doi.org/10.1101/342329
- Multi-Type Birth Death Model: https://doi.org/10.1093/molbev/msw064
2019-02-18: TtB Structured Models
By Nicola Mueller
2019-02-18: TtB Structured Models
- 1,945



