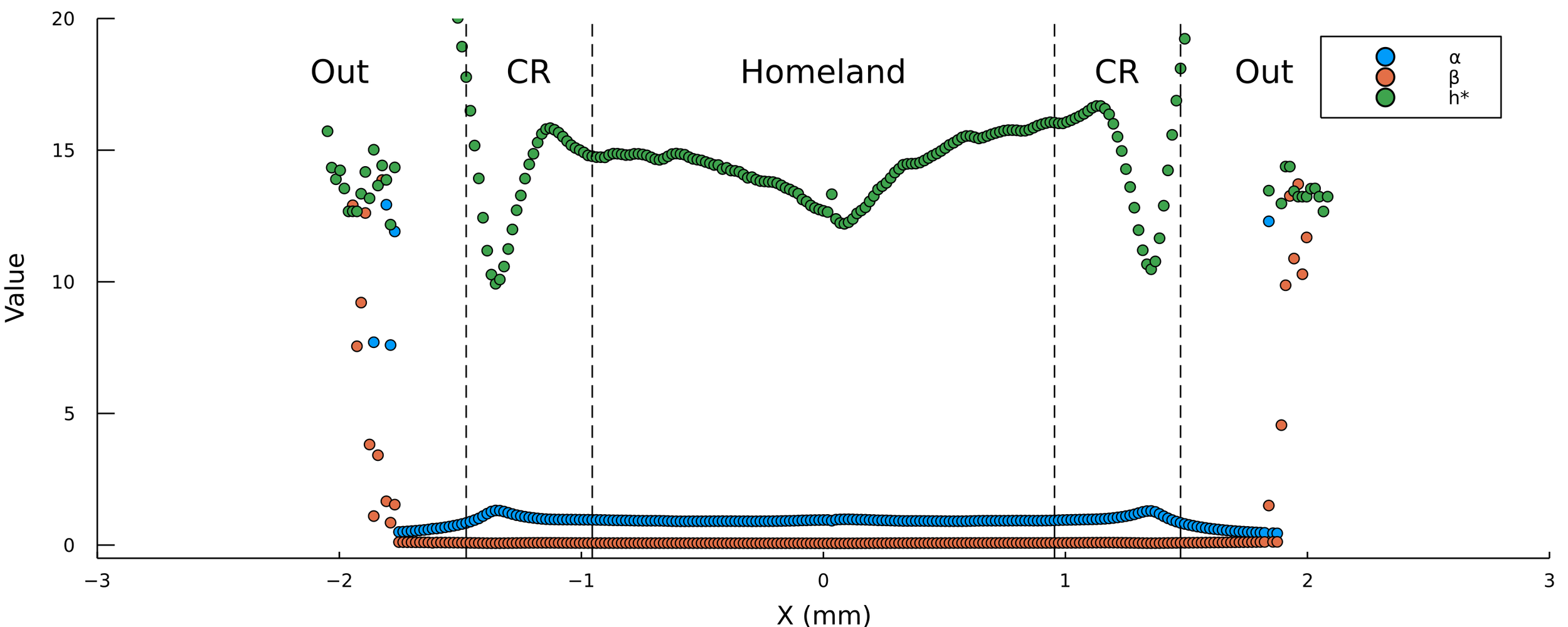
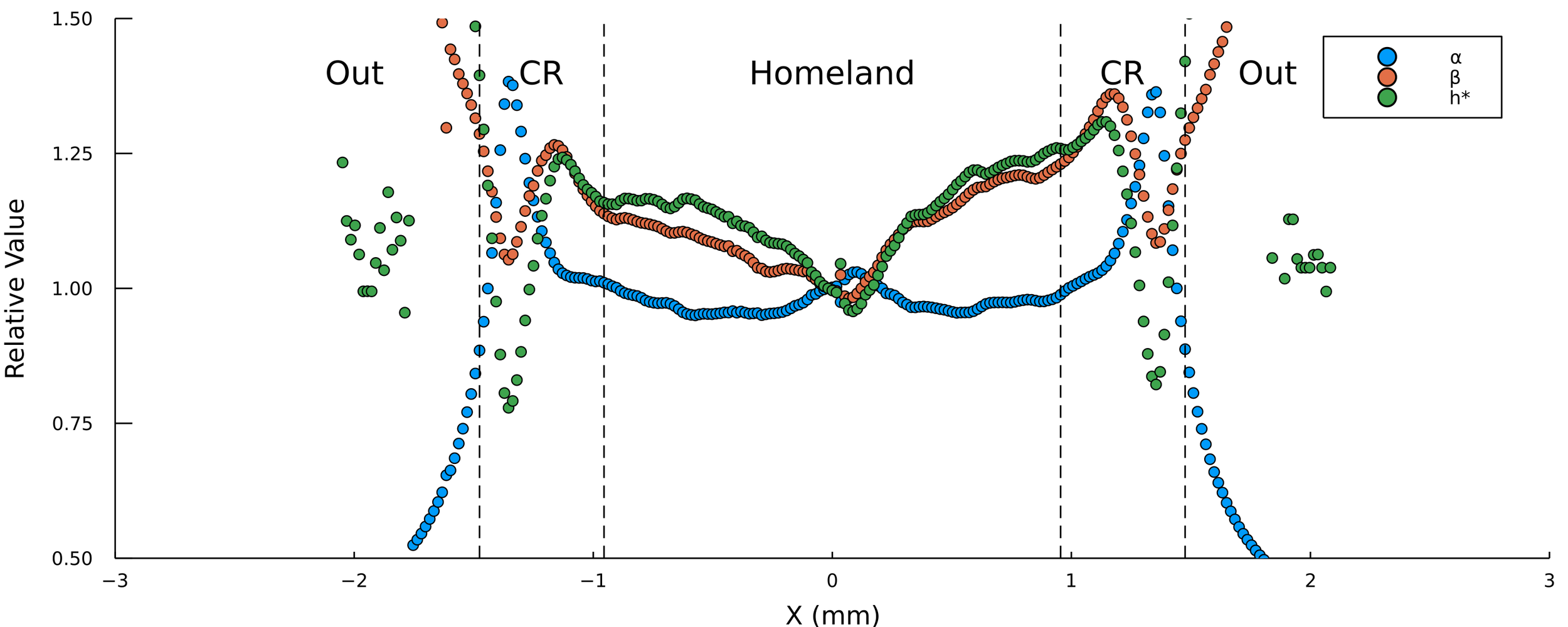
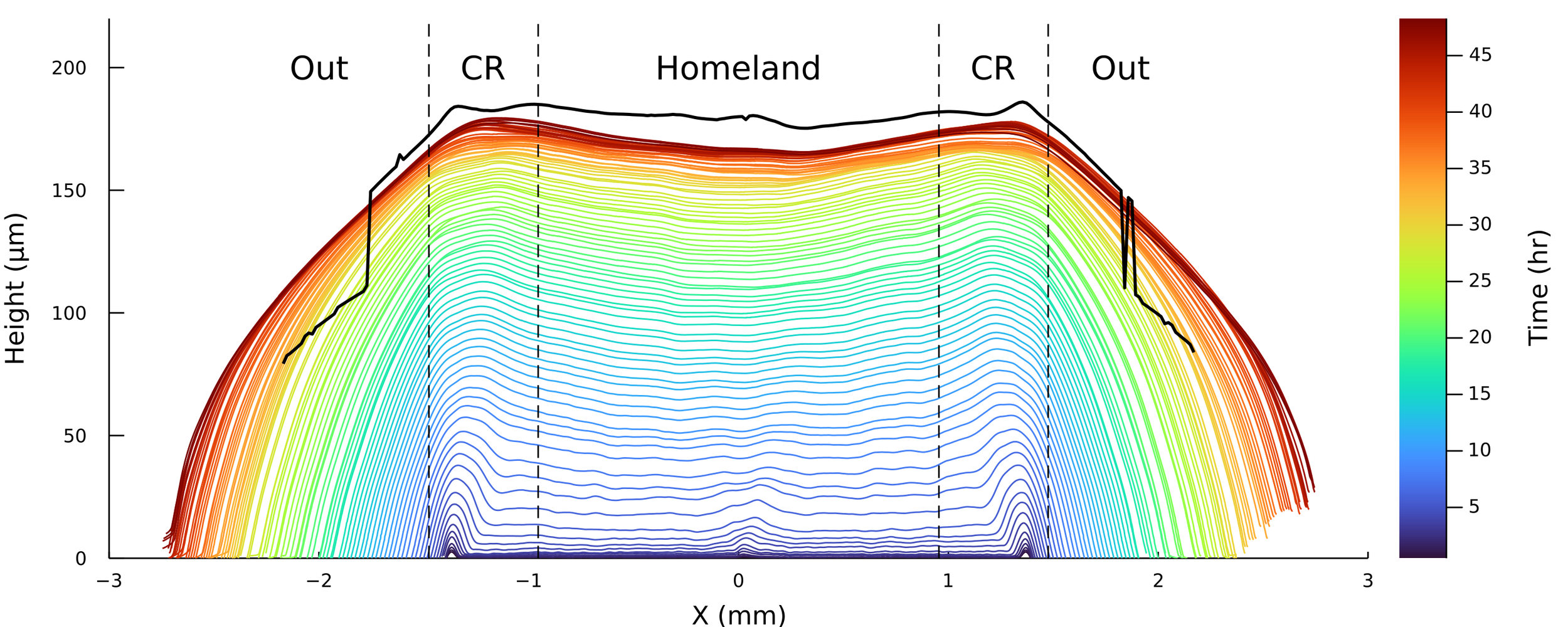
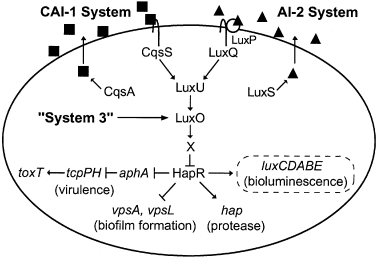
Pablo Bravo PRO
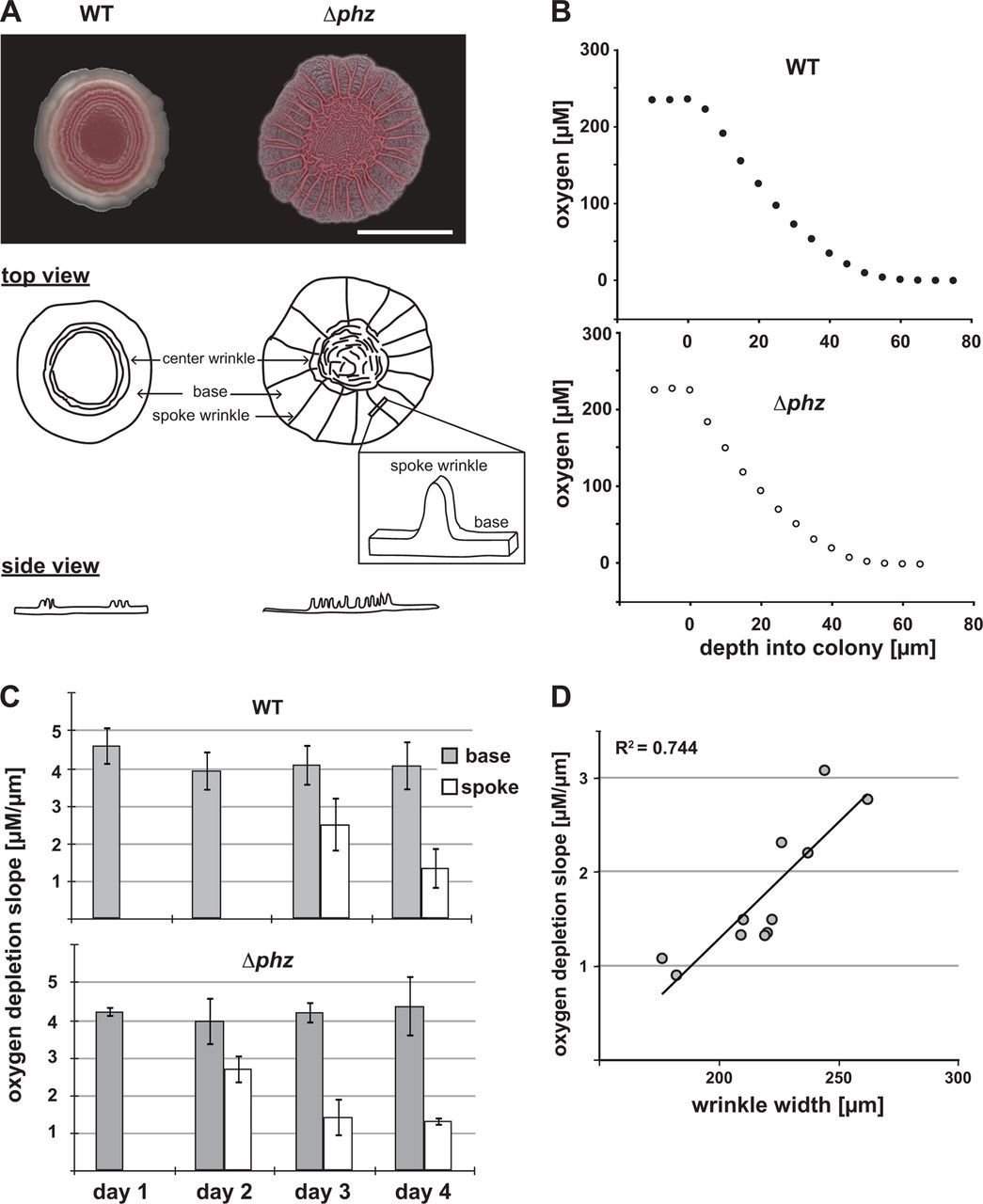
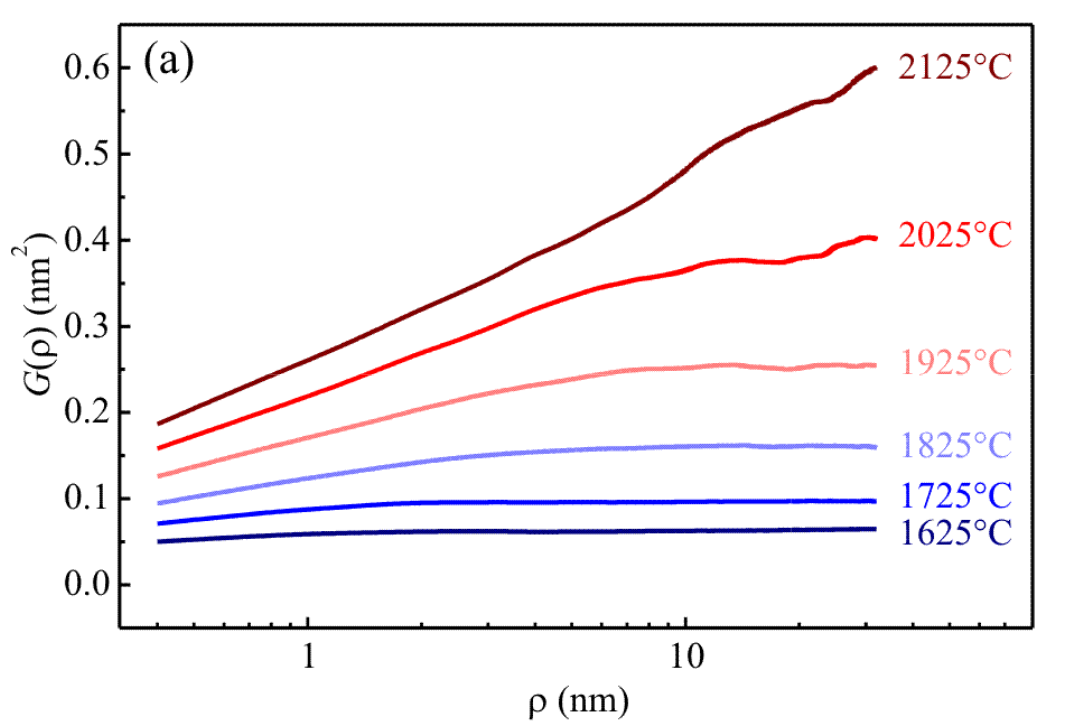
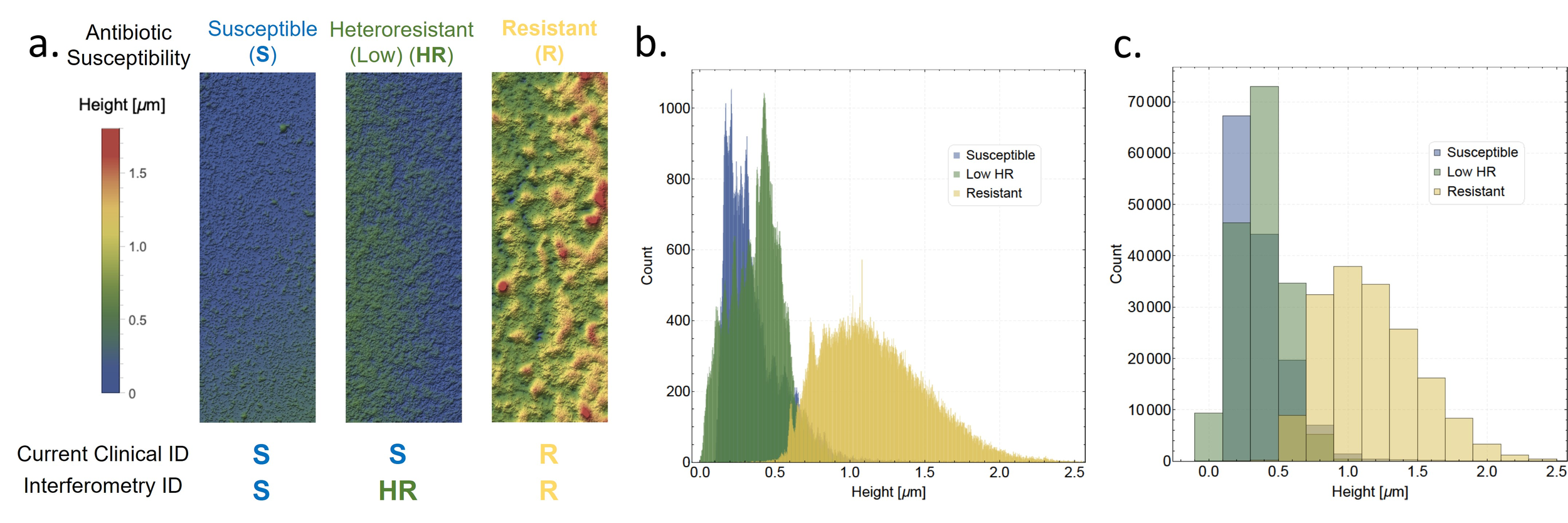
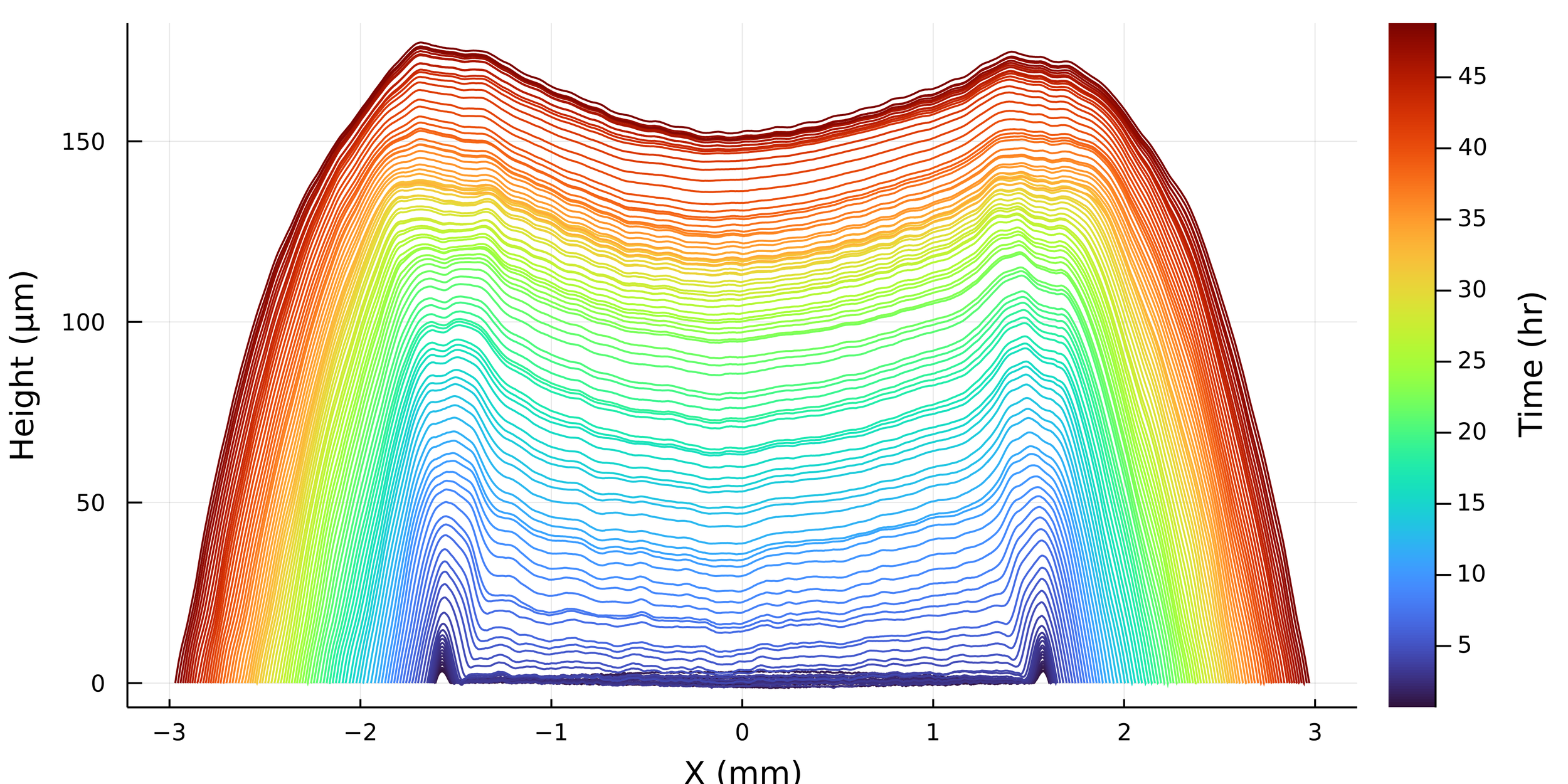
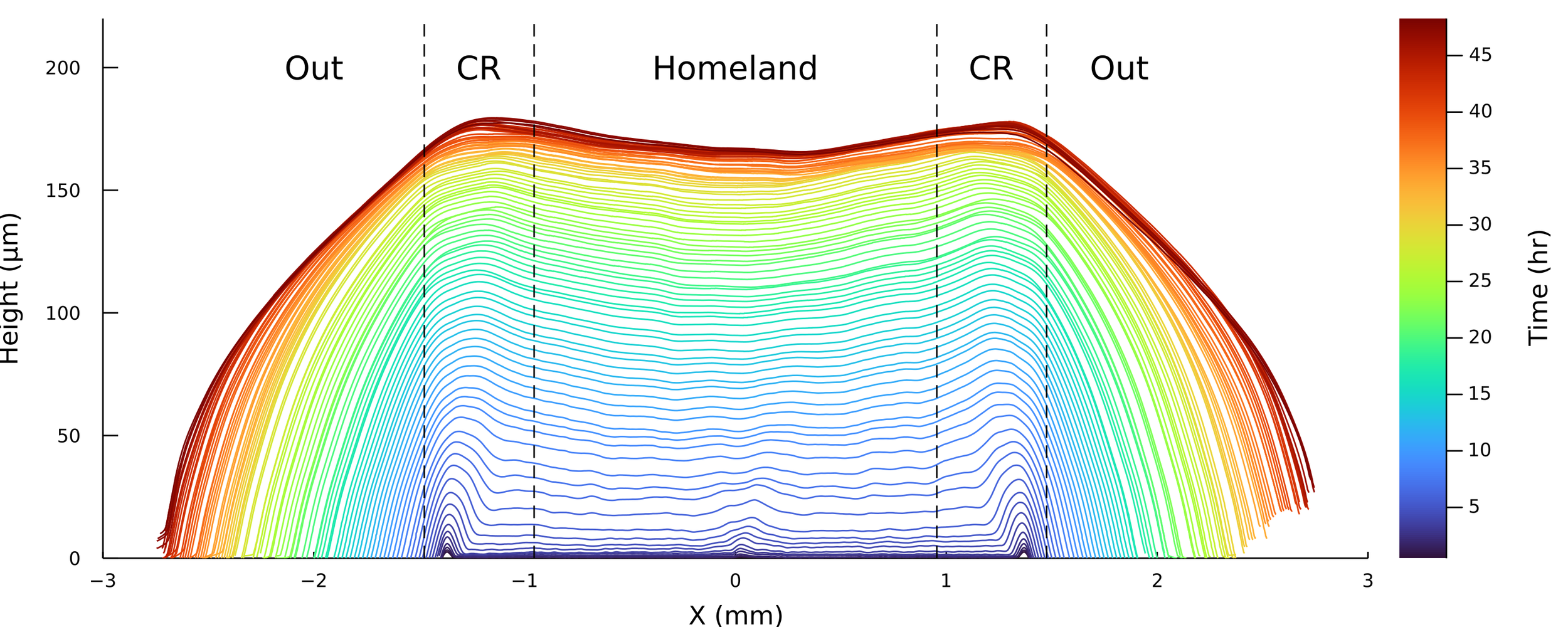
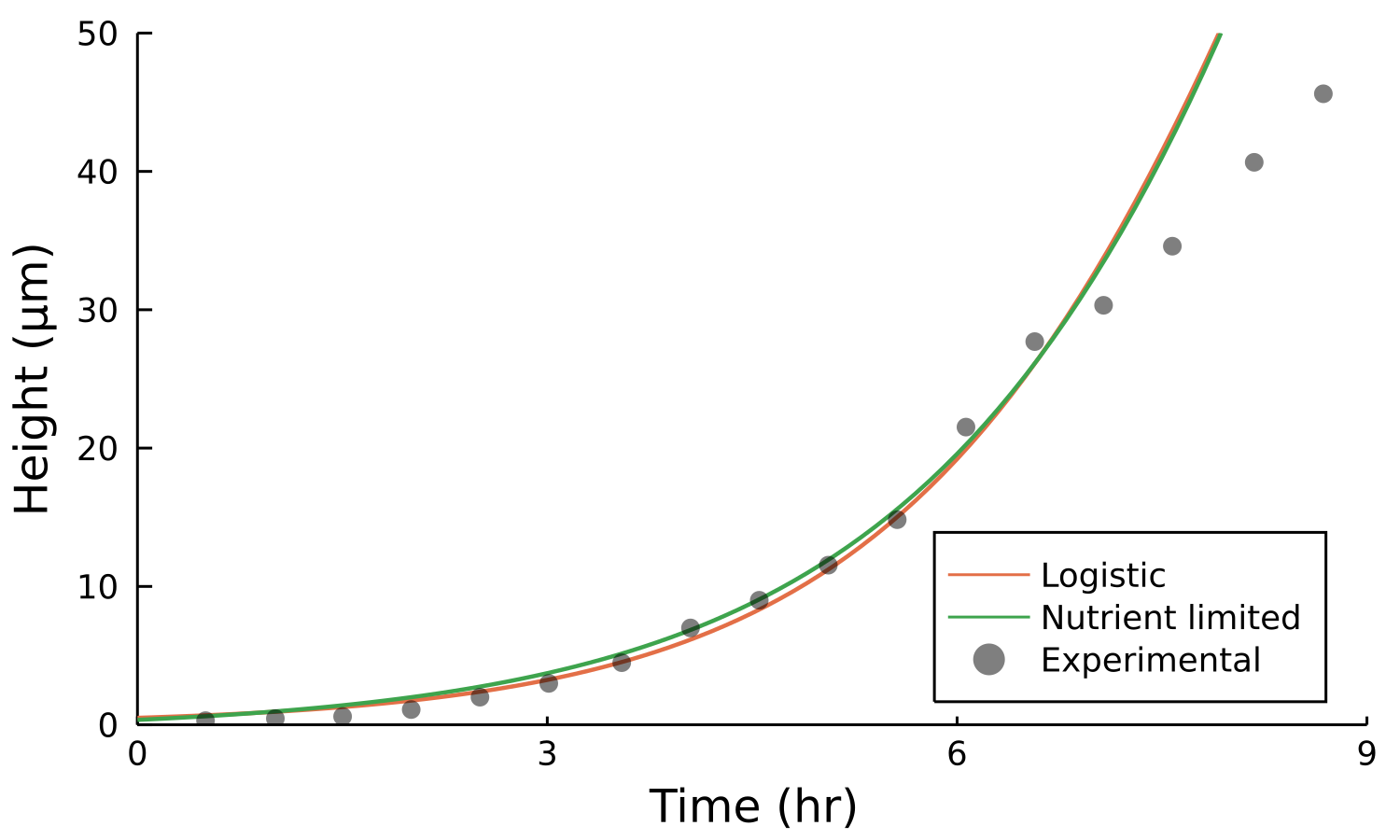
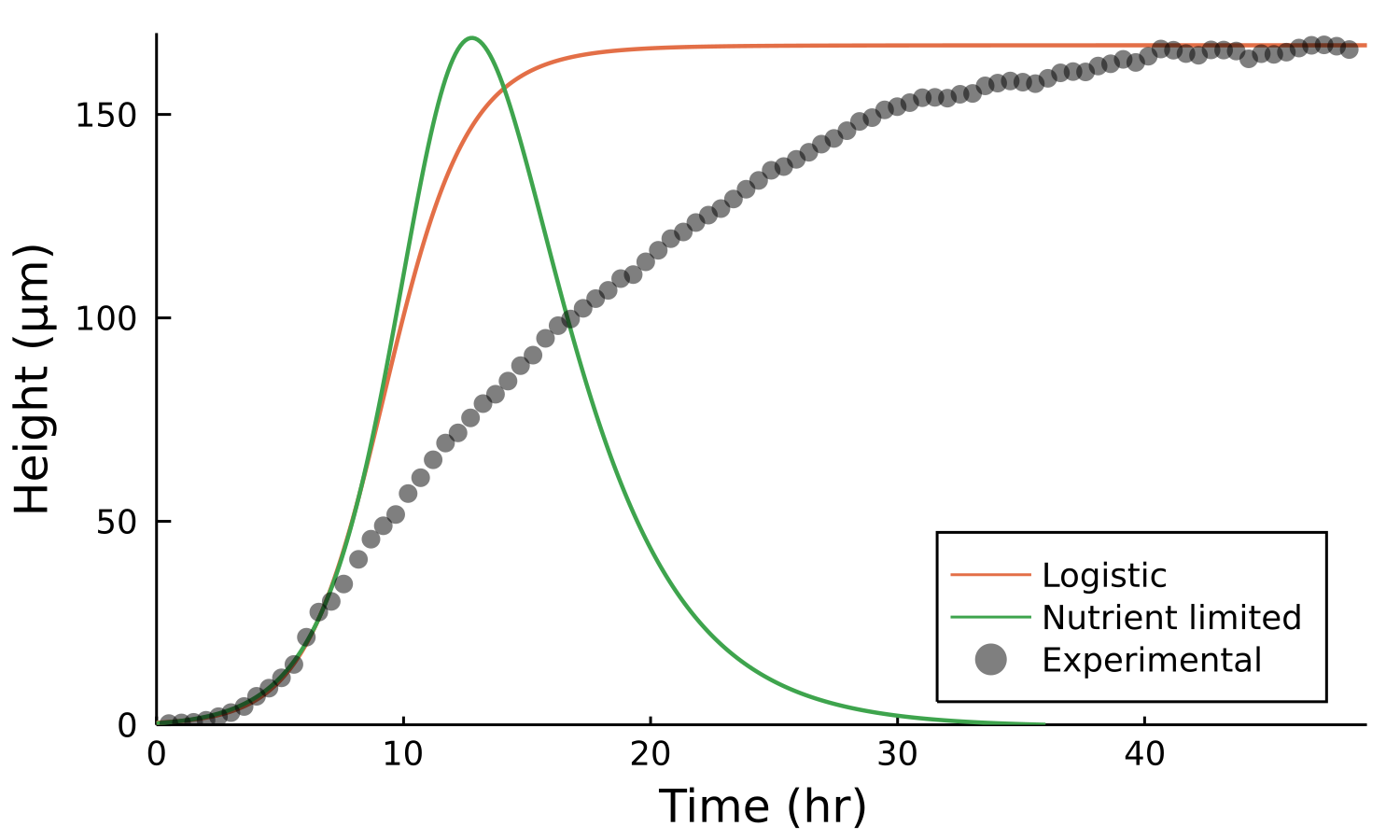
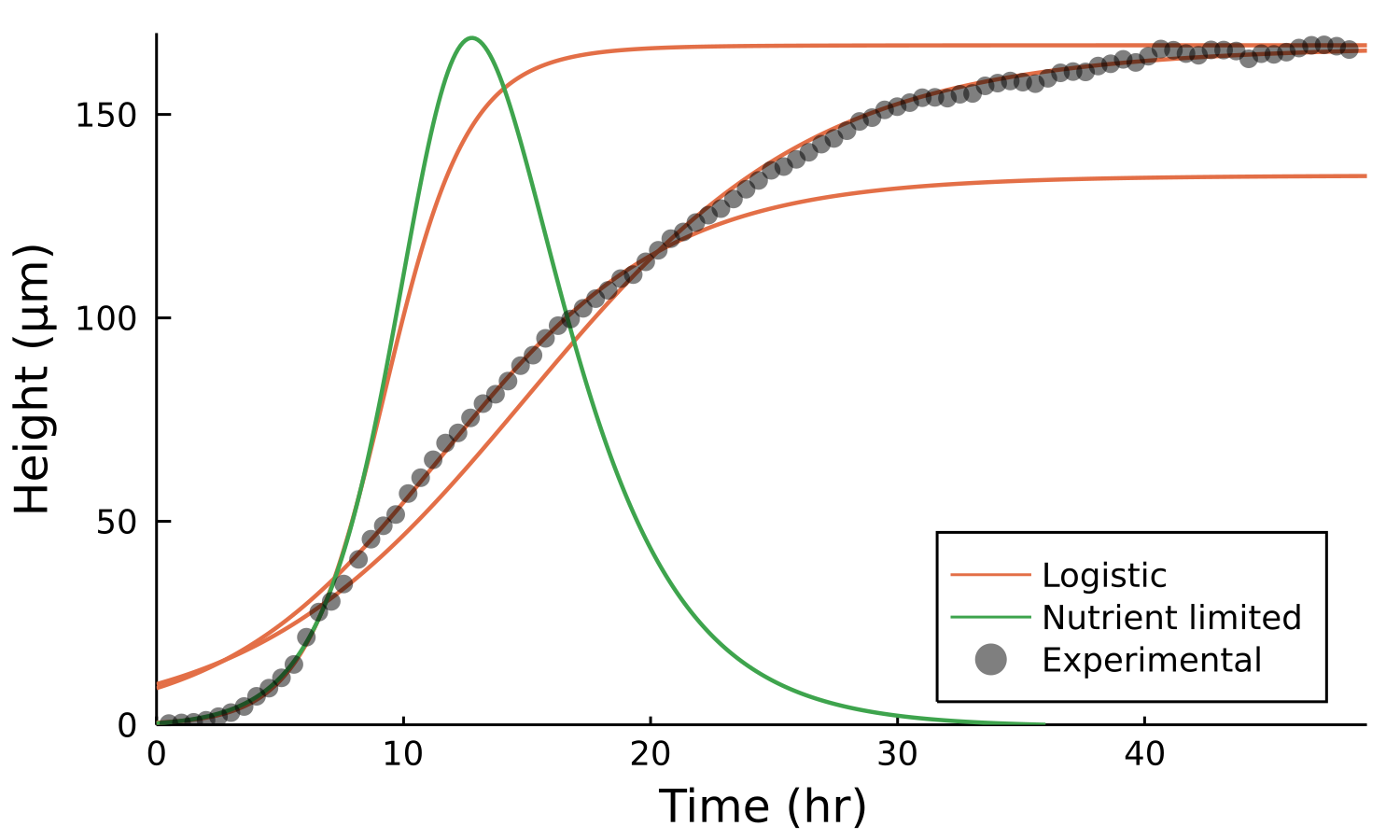
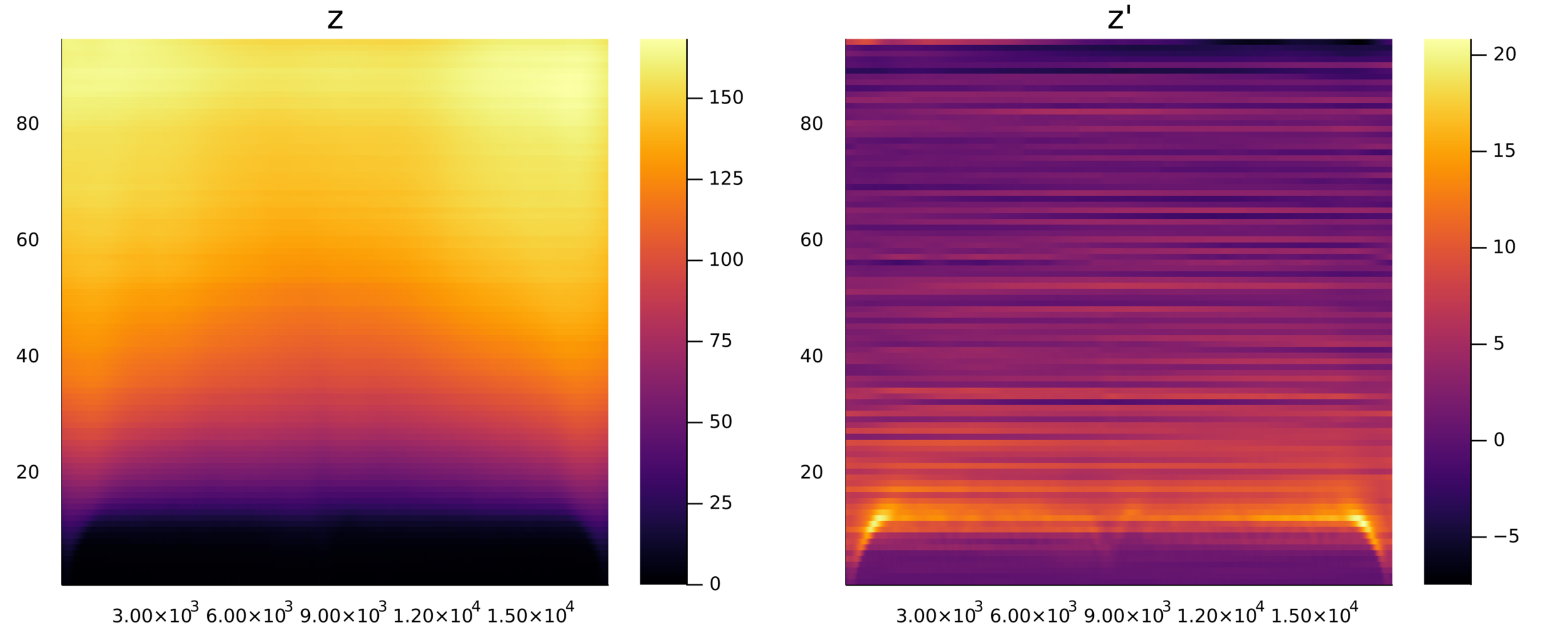
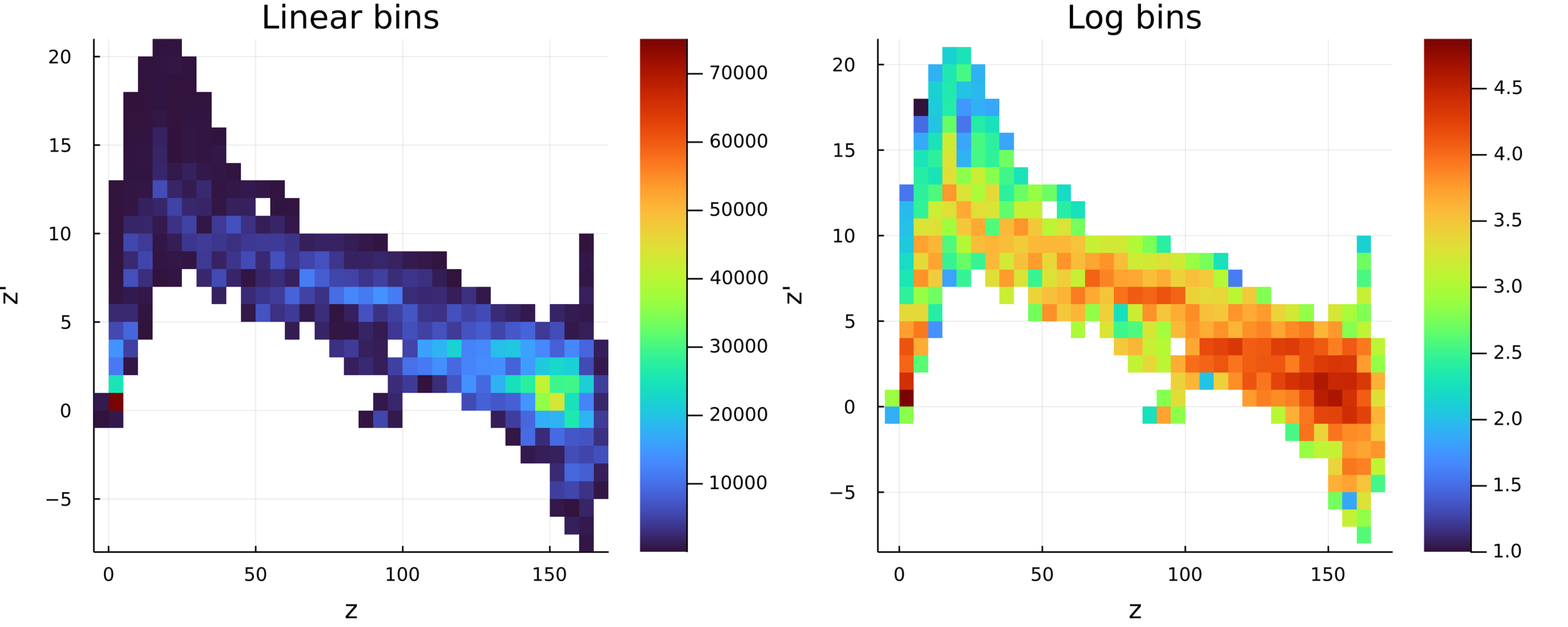
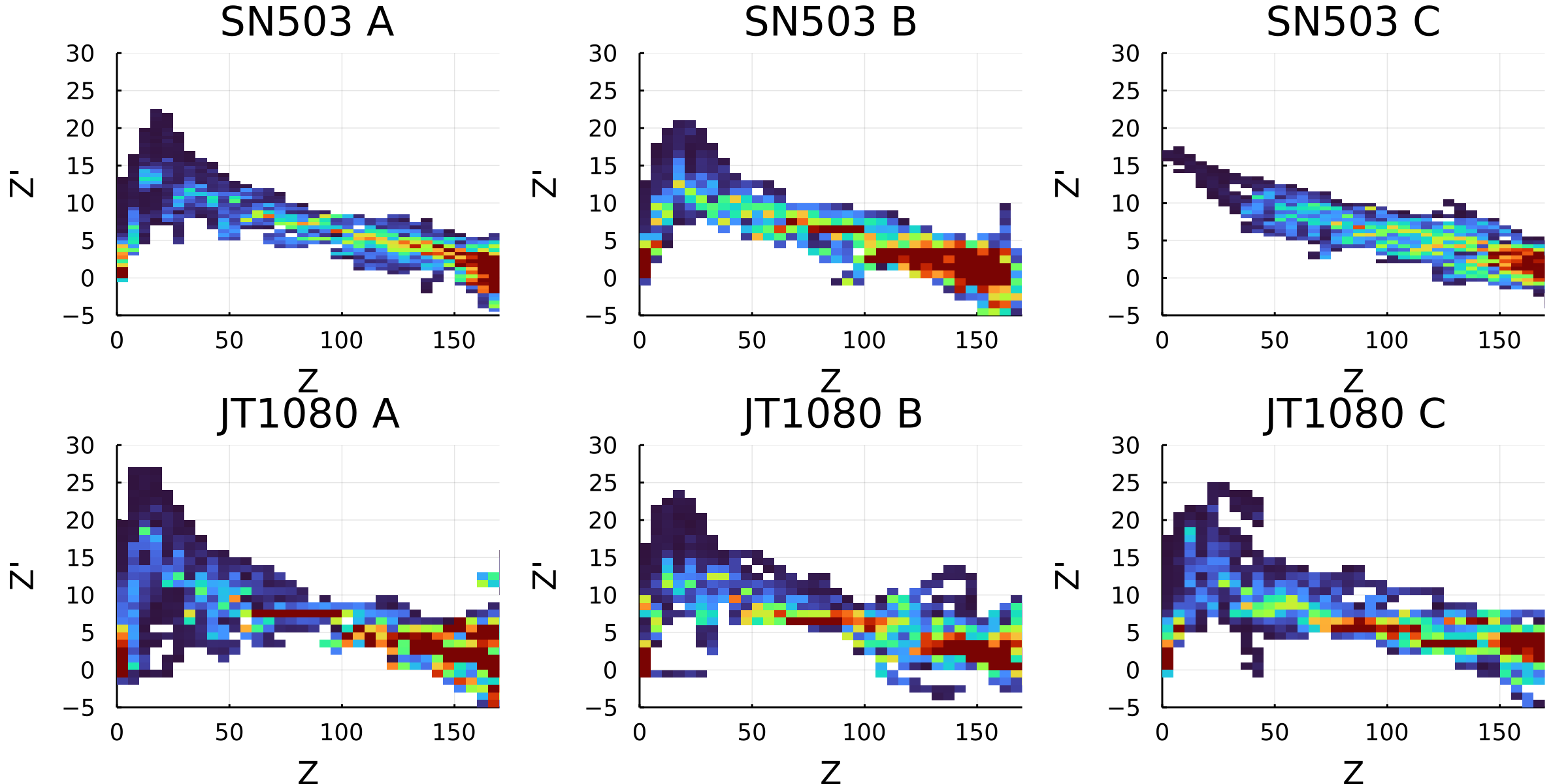
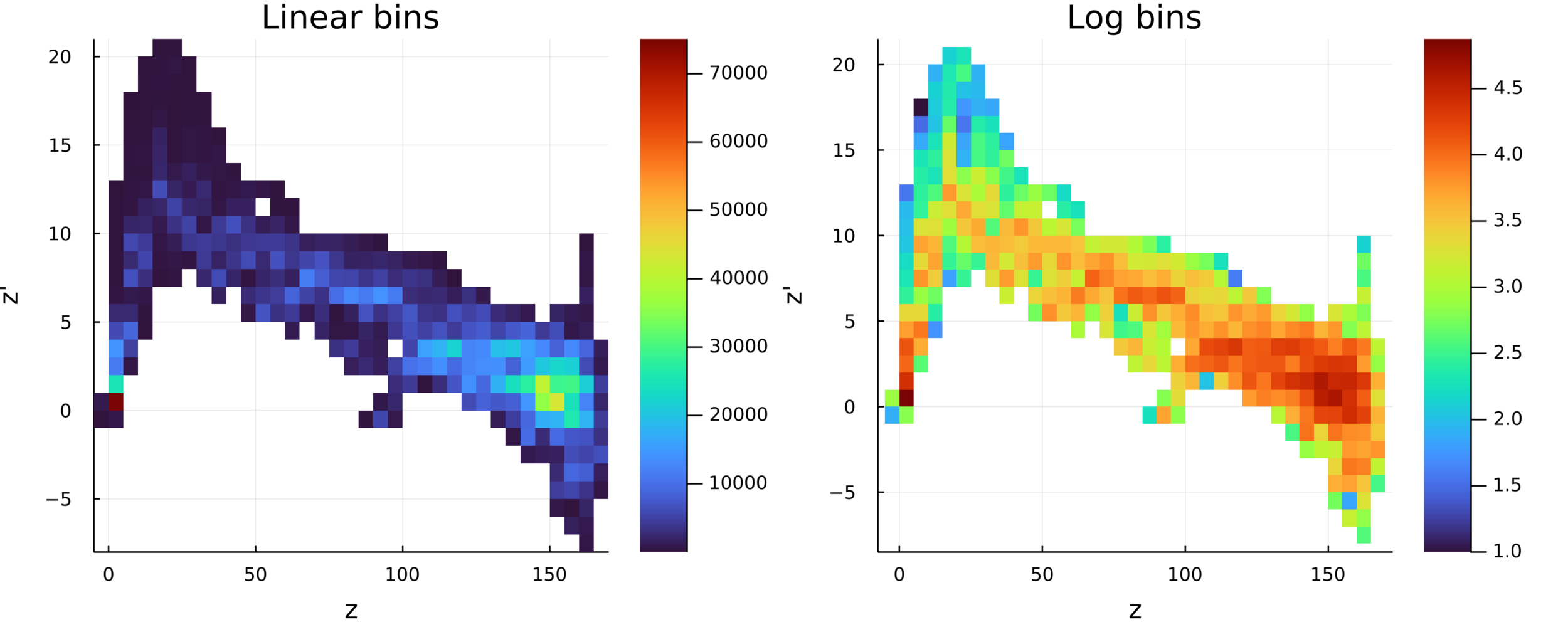
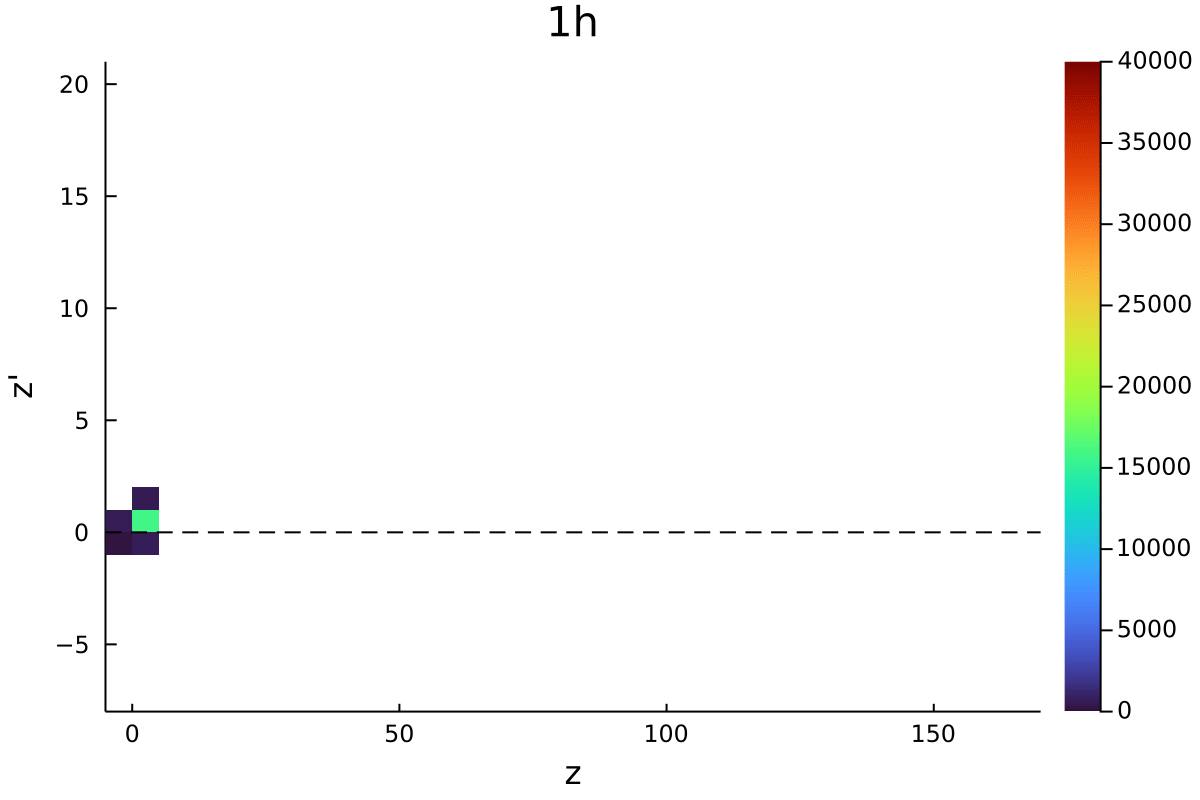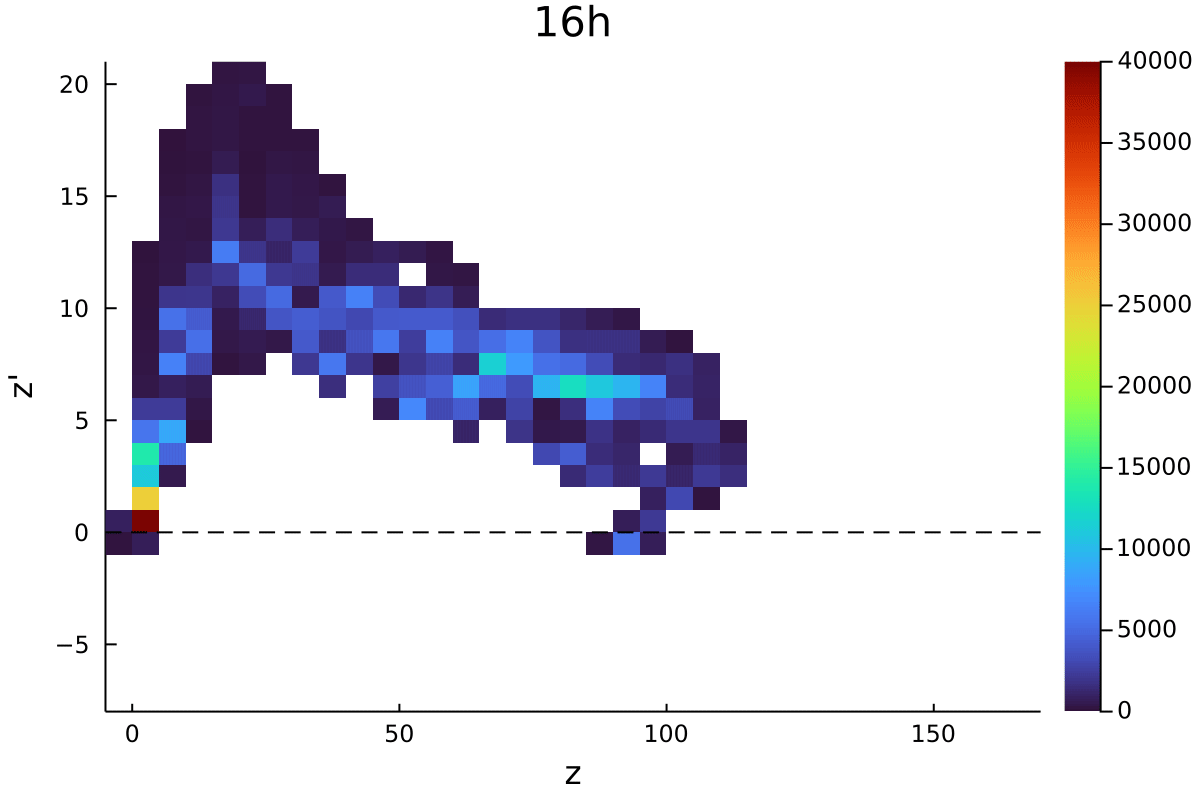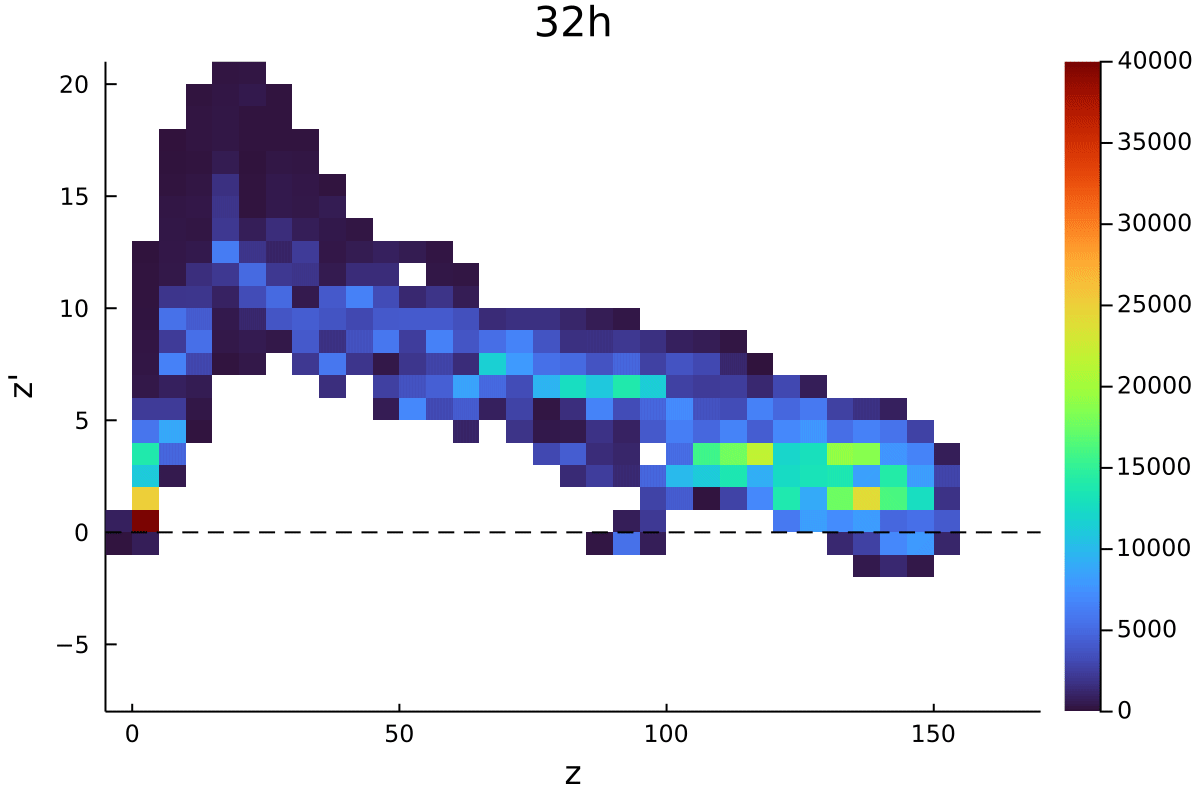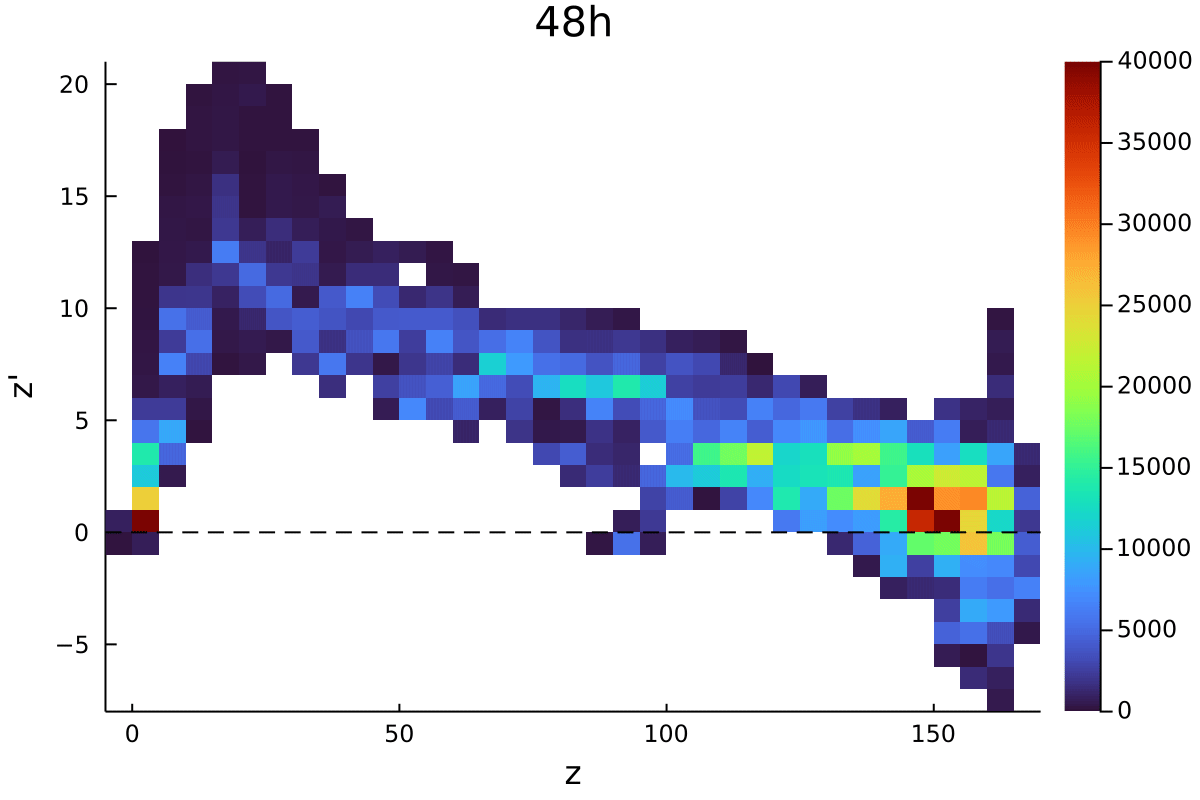
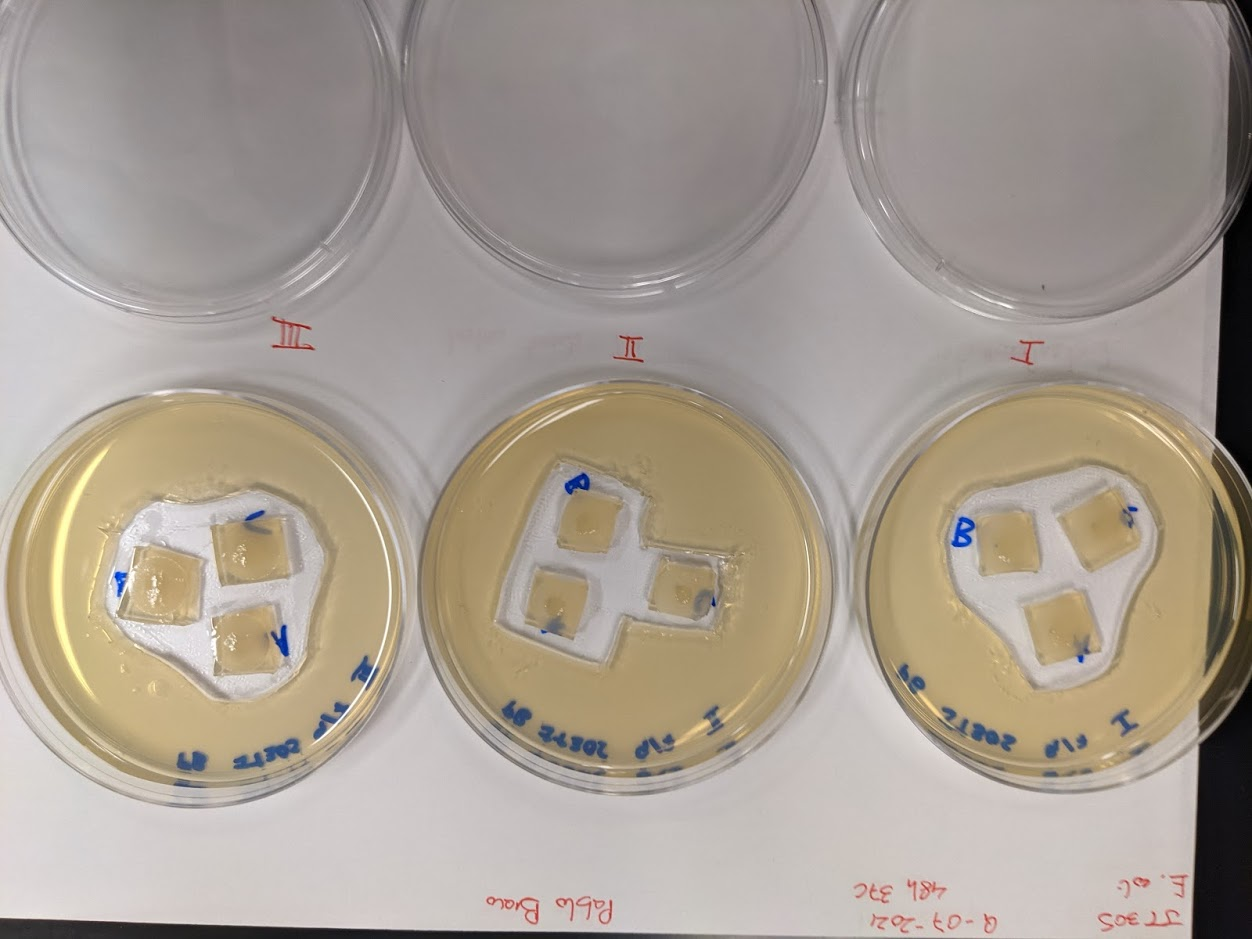
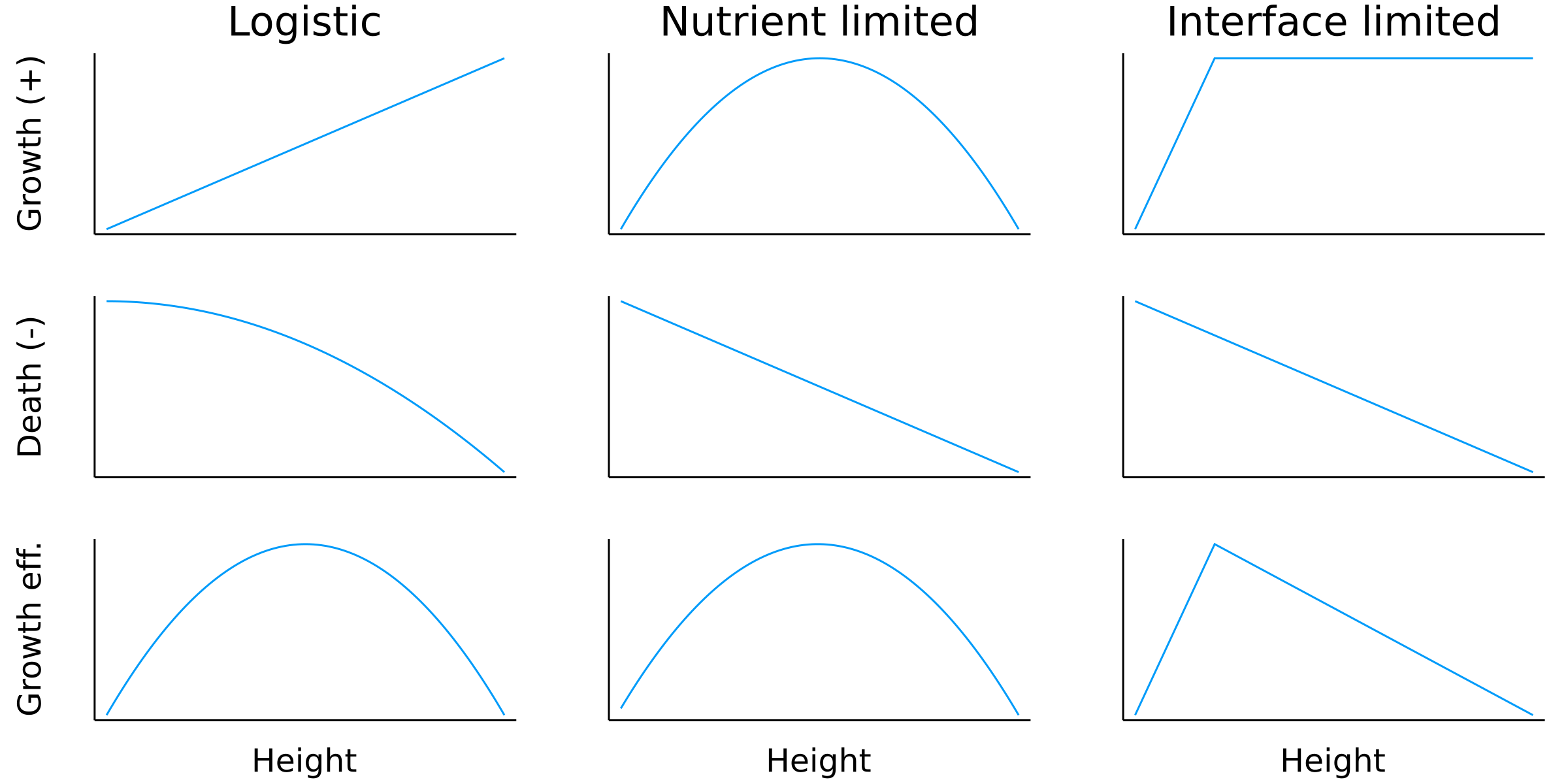
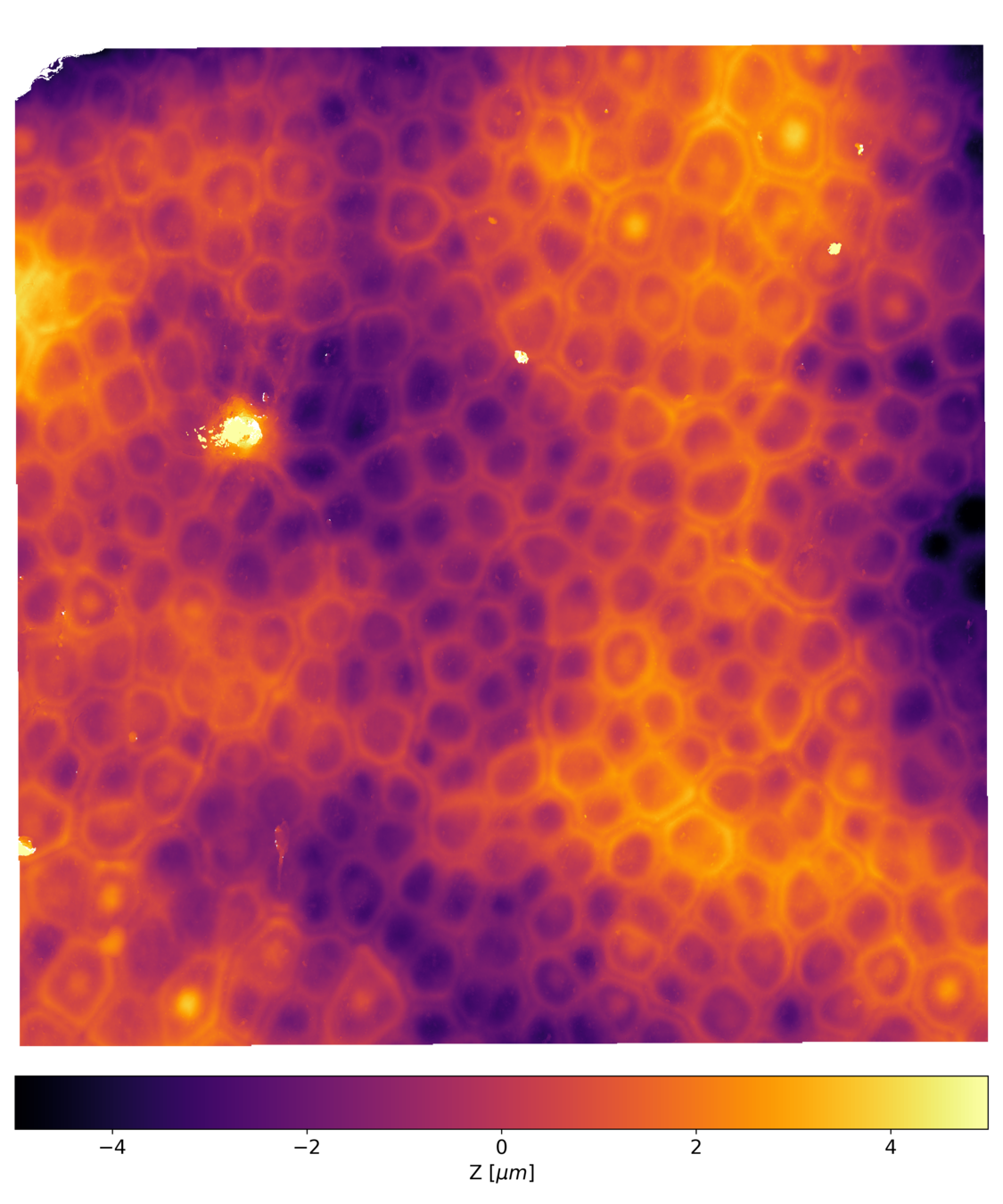
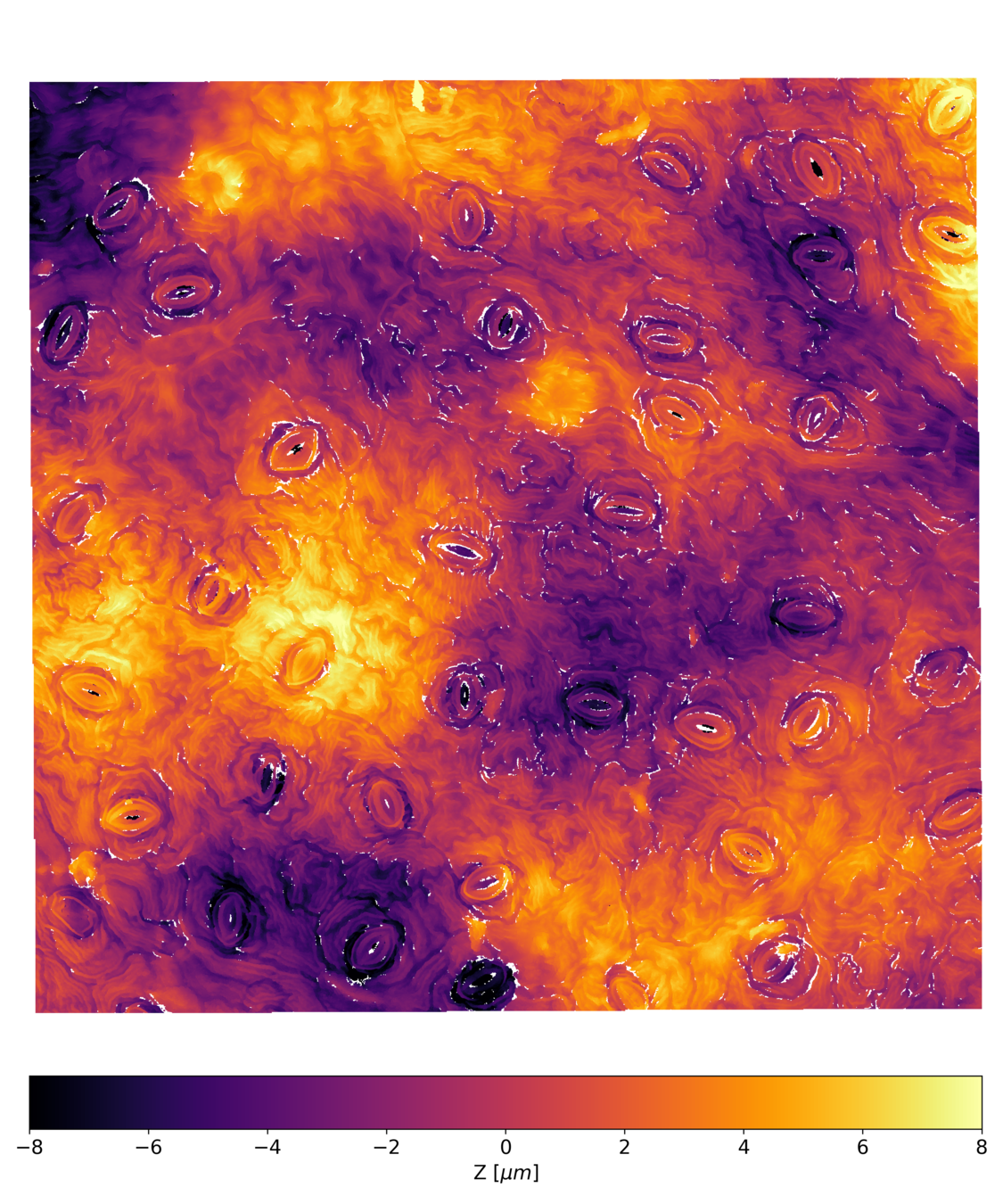
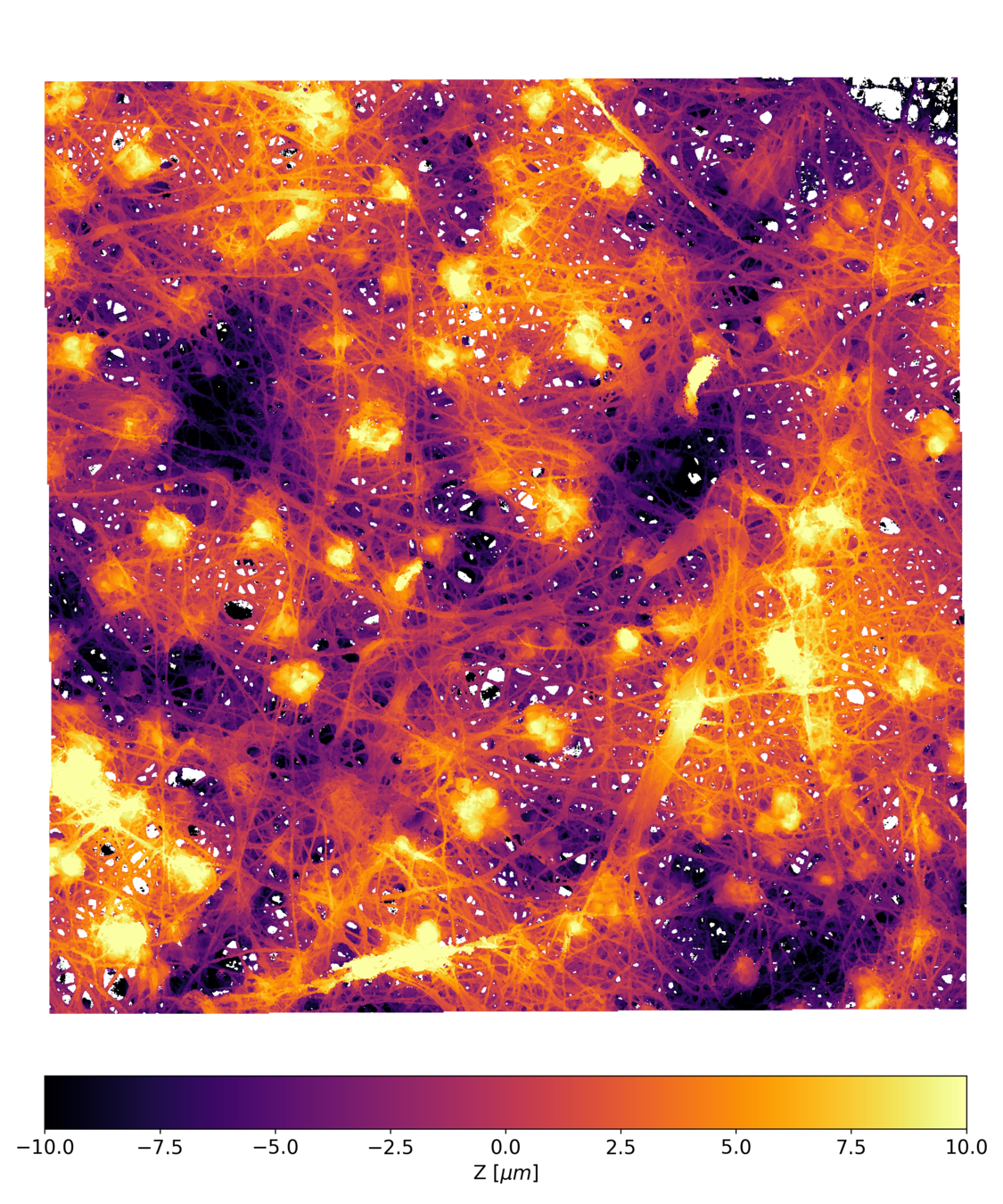
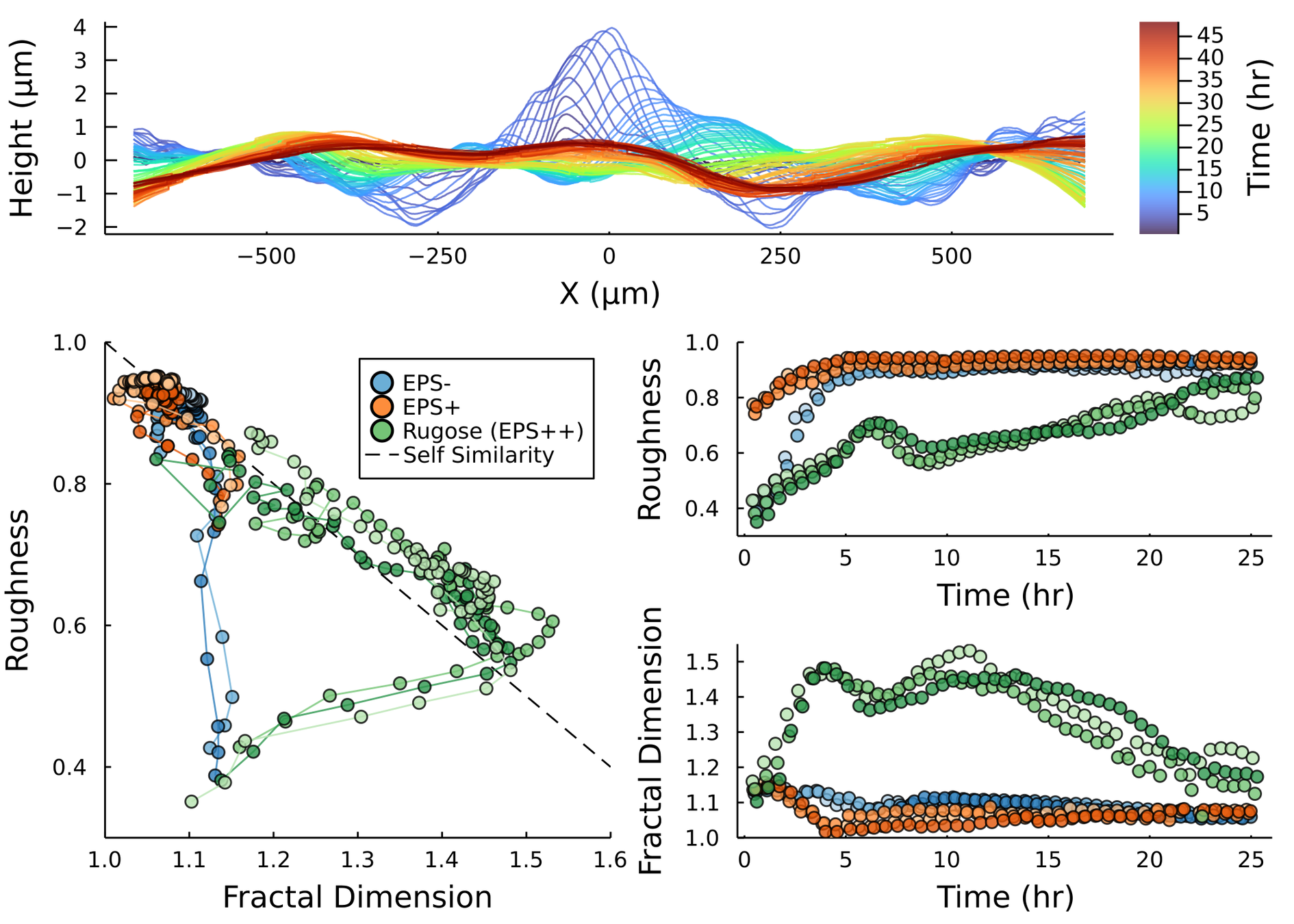
Postdoctoral research at GeorgiaTech. I am interested in biofilms, and how their topography evolves and what we could learn about their development by looking at them. Currently studying how colony size affects resilience towards antibiotic stresses