Enabling the use of gene-by-gene typing methods through a public and centralized service

pedrorvc

Pedro Vila-Cerqueira • Mário Ramirez Lab
Chewie-NS
Bioinformatic Open Days 2020

Is Patient A strain the same as Patient B's?

Patient A


Patient B
Gene-by-Gene Based Typing Methods
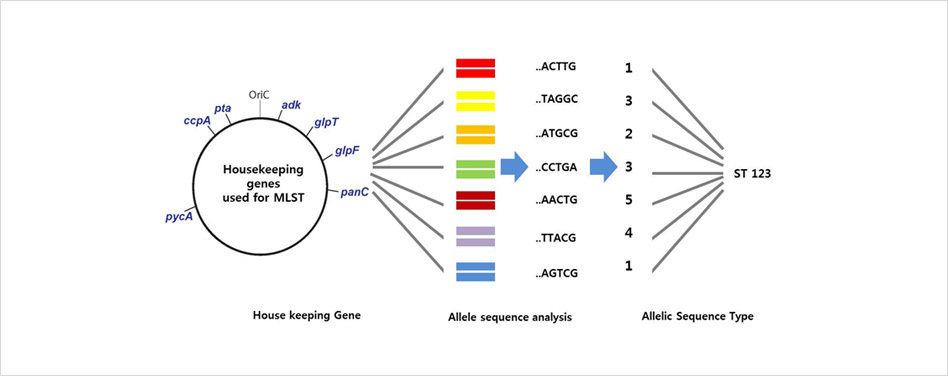
Multilocus Sequence Typing (MLST)
- Defined scheme of typically 7 housekeeping gene fragments.
- Robust, portable and unified method for characterizing isolates at a molecular level.
- Not enough resolution to perform high resolution typing.
-
Whole-Genome Multilocus Sequence Typing (wgMLST)
- Extend MLST to whole-genome level
- Set of genes that are present across a set of genomes representing a species, akin to a pan-genome.
-
Core-Genome Multilocus Sequence Typing (cgMLST)
- Gene-by-gene allelic profiling of core genome genes in a set of same species isolates.
PCR & Sanger Sequecing
Whole Genome Shotgun Sequencing
Gene-by-Gene Based Typing Methods
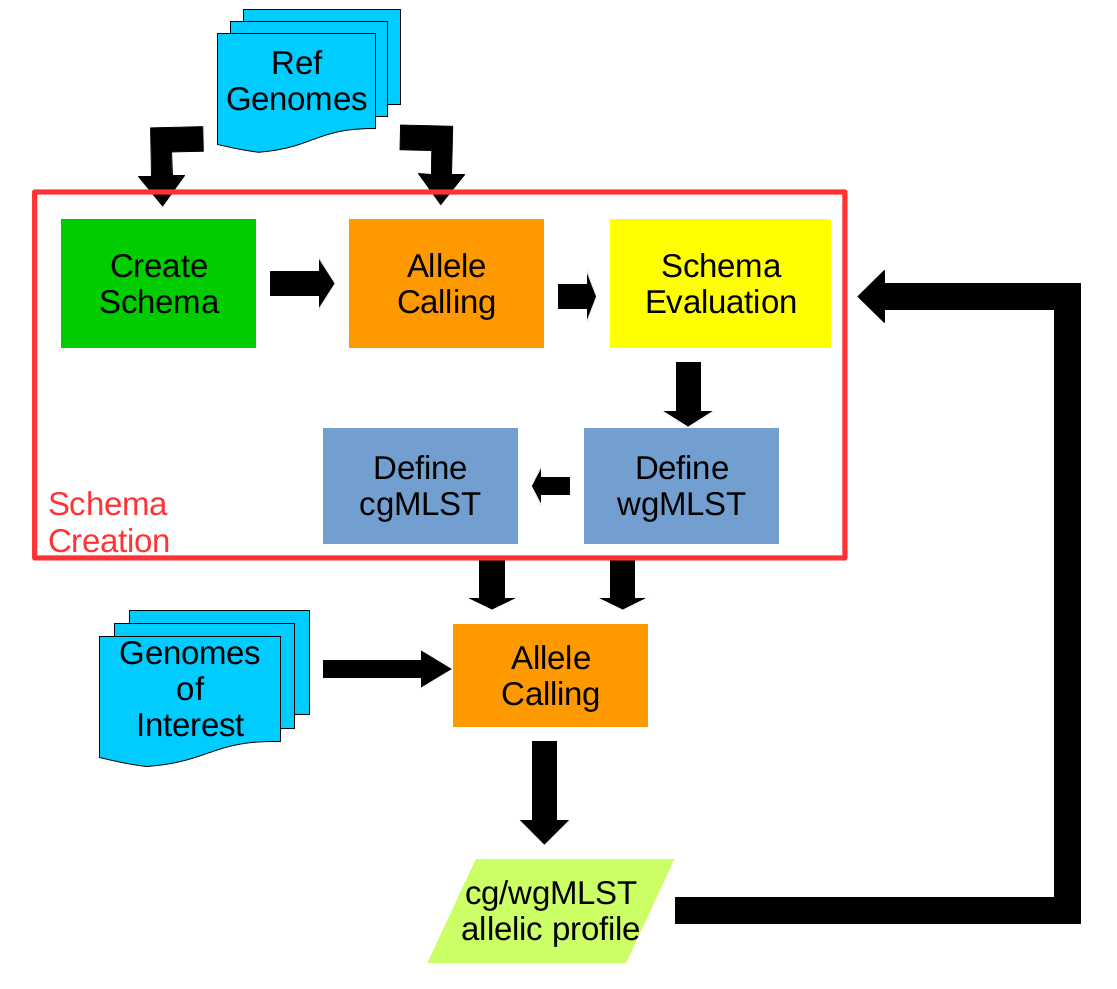
- BLAST Score Ratio (BSR) Based Allele Calling Algorithm.
- Open source solution for the creation of whole genome and core genome MultiLocus Sequence Typing (wg/cgMLST) schemas.
- Performs allele calls on complete or draft genomes resulting from de novo assemblers and determines allelic profiles.
chewBBACA
https://github.com/B-UMMI/chewBBACA
- Profiles
| File | Locus 1 | Locus 2 |
|---|---|---|
| genome1.fasta | Allele 2 | Allele 5 |
| genome2.fasta | Allele 6 | Allele 9 |
- Profile Visualization
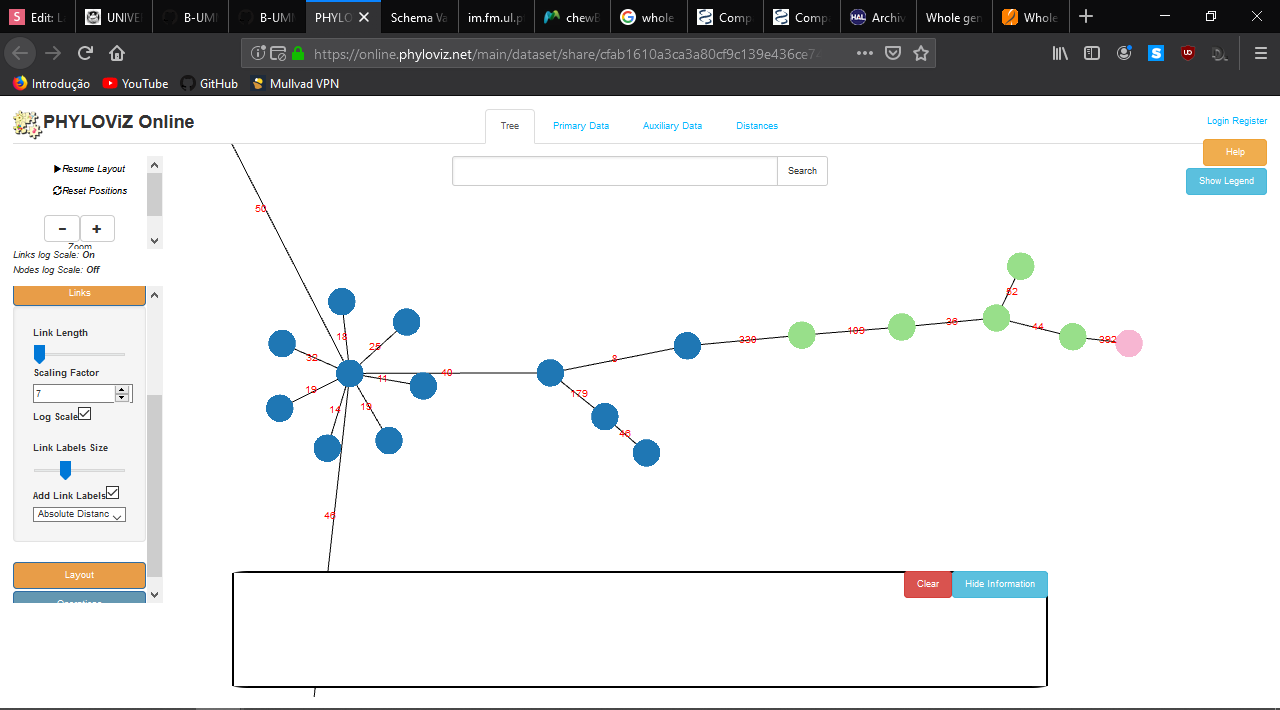
chewBBACA
https://online.phyloviz.net
Core-Genome Multilocus Sequence Typing (cgMLST)
Is there an outbreak?









What data to share?
- Allelic profiles?
- Raw data?
Strict privacy laws may prevent the users from sharing raw data. Unpublished data is also a matter of concern.



- Schemas?
Schemas can be created with different parameters, requiring the users to share their configurations.
Profiles are generated based on the schema, which may contain private data, making it hard to share and obtain the same results.
What data to share?
-
Provide a public and centralized web service enabling users to:
- Download the necessary data for the wg/cgMLST schemas;
- Query/submit results to the database.
- Integration with chewBBACA allows users to perform analyses in their local machines.
Chewie-NS: a nomenclature server
- Share and compare allelic profiles within the community at a global level.




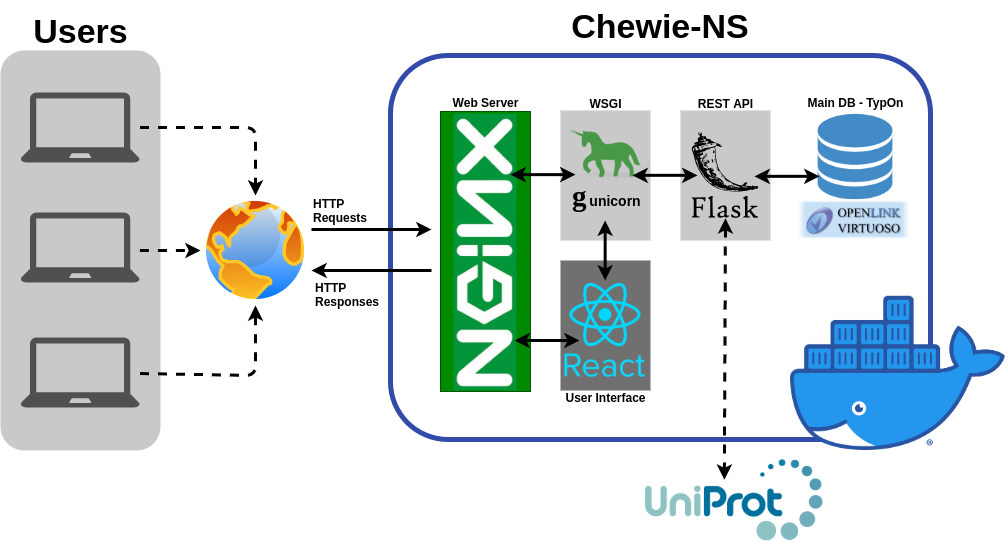
Chewie-NS: a nomenclature server
Chewie-NS: a nomenclature server
- HTTP server and reverse proxy and load balancer.
- WSGI server that handles HTTP requests and routes them to any Python application that is WSGI-compliant, such as Flask.
- Database management system for RDF data.
- OWL ontology for describing microbial typing.
- https://doi.org/10.1186/2041-1480-5-43
NGINX:
Gunicorn:
Virtuoso Triple Store:
TypOn:
- WSGI: Web Server Gateway Interface
- RDF: Resource Description Framework
- OWL: Web Ontology Language




Git, Docker and Docker-compose to run multi-container Docker applications.
apt-get install gitapt-install docker-cecurl -L "https://github.com/docker/compose/releases/download/1.25.3/docker-compose-$(uname -s)-$(uname -m)" -o /usr/local/bin/docker-composegit clone https://github.com/B-UMMI/Chewie-NS.gitClone the repo
Deploy Chewie-NS

docker-compose up

Docker Compose
Chewie-NS: a nomenclature server
Chewie-NS: a nomenclature server
DEMO API

Chewie-NS: a nomenclature server
DEMO UI

- Chewie-NS aims to provide all the necessary functions to download and upload schemas and submit alleles to the server in a flexible way.
- Users can work with their data locally, circumventing possible concerns over data privacy in sharing data.
- Possibility to synchronize schemas without sending any data to the server, maintaining their novel alleles private.
- If they so wish, users can submit their novel results to the web service.
Chewie-NS: a nomenclature server
https://github.com/B-UMMI/Chewie-NS

Special thanks to Rafael Mamede, Inês Mendes, Mickael Silva, João André Carriço and Mário Ramirez.
https://github.com/B-UMMI/chewBBACA
This work was partially supported by the following projects: the ONEIDA project (LISBOA-01-0145-FEDER-016417) co-funded by FEEI – ‘Fundos Europeus Estruturais e de Investimento’ from ‘Programa Operacional Regional Lisboa 2020’ and by national funds from FCT – ‘Fundação para a Ciência e a Tecnologia’, BacGenTrack (TUBITAK/0004/2014) [FCT/Scientific and Technological Research Council of Turkey (Türkiye Bilimsel ve Teknolojik Araşrrma Kurumu, TÜBİTAK)] and LISBOA-01-0145-FEDER-007391, project cofunded by FEDER, through POR Lisboa 2020 - Programa Operacional Regional de Lisboa, PORTUGAL 2020, and Fundação para a Ciência e a Tecnologia.



Thank you for your attention!
https://github.com/B-UMMI/Chewie-NS_tutorial
Chewie-NS:
By Pedro Cerqueira
Chewie-NS:
CBBS and BOD'20
- 726



