Welcome to Data Mining
With :

BOUDALI Soheib
Etudiant M2 AI USTHB
Fondateur de Blactus Technologies
Powered by : Blactus Technologies
What's DM !
DM USAGE
HOW !
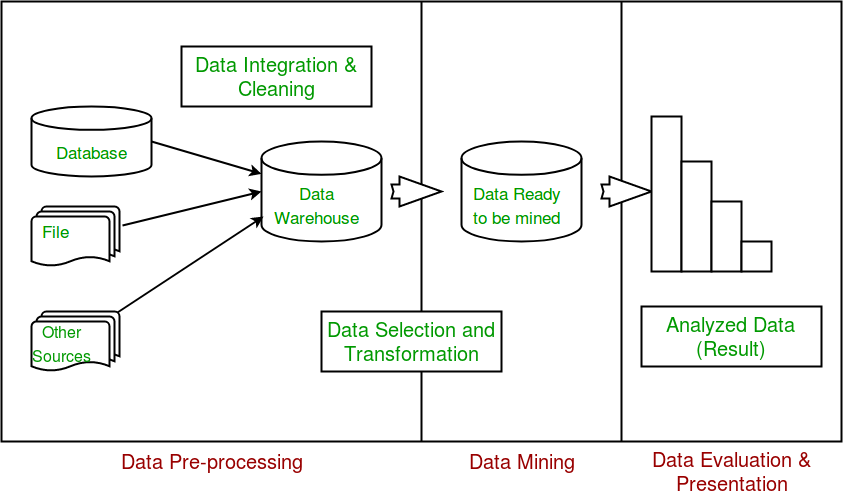
Data exploration
DATASET Manipulation
Data set opening , reading and printing
file=open("dataset_name","r",encoding="utf-8")
dataset=file.readlines()
for line in dataset:
print(line)Extract attributes type
- float
- string
- ...
Explore descriptive characteristics of the data
- Symmetry study :
Mean
Mode
Median
Min
Max
import numpy as np
mean=np.mean(array)
import numpy as np
mean=np.median(array)
from scipy import stats
mode =stats.mode(array)
Min= min(array)
Max= max(array)
import pickle
import numpy as np
from scipy import stats
pkl_file = open('heart.pkl', 'rb')
dataset= pickle.load(pkl_file)
pkl_file.close()
column=3
array=[]
for i in range(0,len(dataset)):
array.append(dataset[i][column])
mean=np.mean(array)
median= np.median(array)
mode =stats.mode(array)
Min= min(array)
Max= max(array)
print("la moyenne est :"+str(mean)+"\n"+"la mediane est : "+str(median)+"\n"+" le mode est: "+str(mode)+"\n")
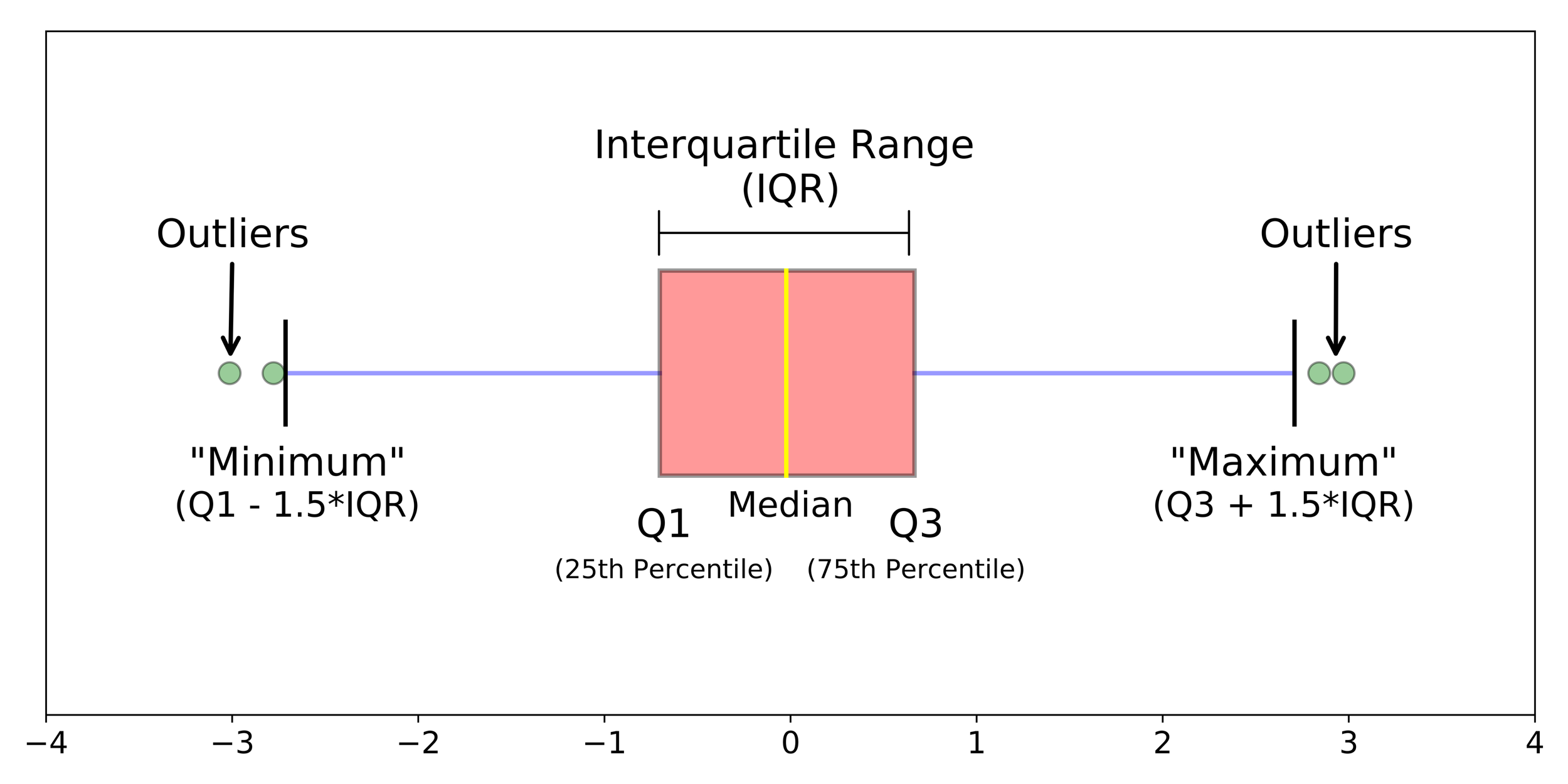
Data visualisation
Boxplot
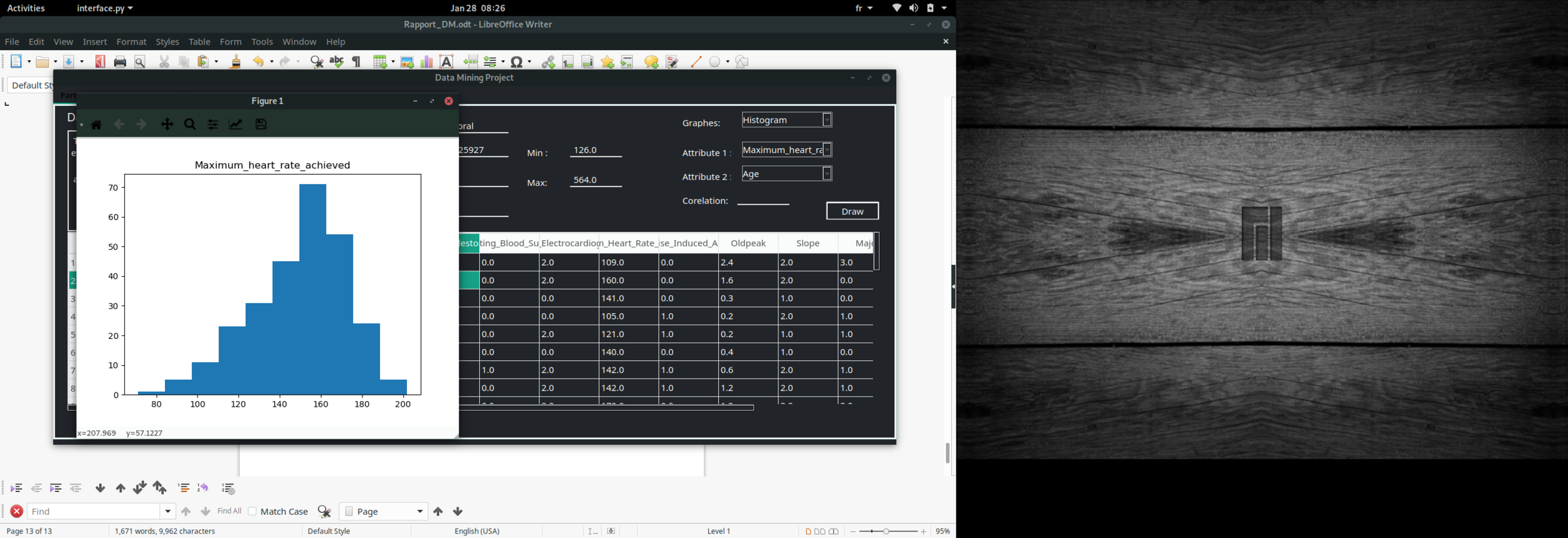
Data visualisation
Histogram
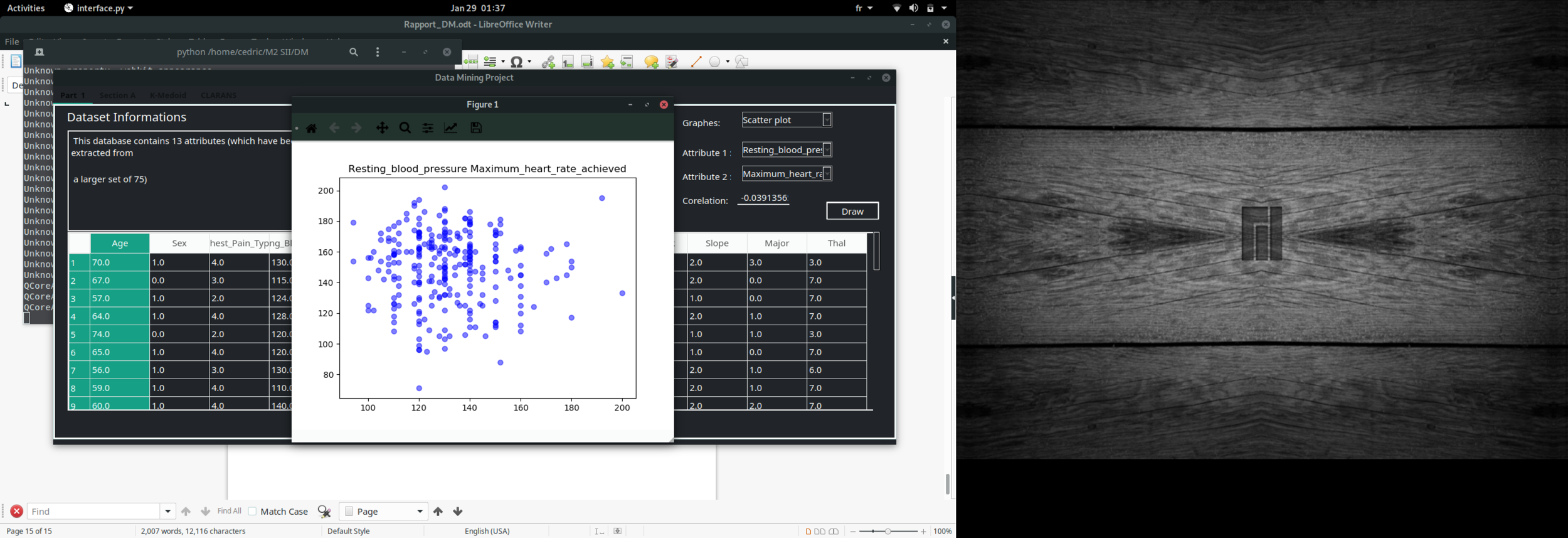
Data visualisation
Scatter plot
import pickle
import numpy as np
import matplotlib.mlab as mlab
import matplotlib.pyplot as plot
def boxplot(Attribute):
pkl_file = open('heart.pkl', 'rb')
dataset= pickle.load(pkl_file)
pkl_file.close()
data=[]
Attributes_names=['Age','Sex','Chest','Resting_blood_pressure','Serum_cholestoral','Fasting_blood_sugar','Resting_electrocardiographic_results','Maximum_heart_rate_achieved','Exercise_induced_angina','Oldpeak','Slope','Number_of_major_vessels','Thal']
for i in range(0,len(Attributes_names)):
if Attributes_names[i]==Attribute:
for instance in dataset:
data.append(float(instance[i]))
fig1, ax1 = plot.subplots()
ax1.set_title(Attribute)
ax1.boxplot(data)
plot.show()
def hist(Attribute):
pkl_file = open('heart.pkl', 'rb')
dataset= pickle.load(pkl_file)
pkl_file.close()
data=[]
Attributes_names=['Age','Sex','Chest','Resting_blood_pressure','Serum_cholestoral','Fasting_blood_sugar','Resting_electrocardiographic_results','Maximum_heart_rate_achieved','Exercise_induced_angina','Oldpeak','Slope','Number_of_major_vessels','Thal']
for i in range(0,len(Attributes_names)):
if Attributes_names[i]==Attribute:
for instance in dataset:
data.append(float(instance[i]))
fig1, ax1 = plot.subplots()
ax1.set_title(Attribute)
ax1.hist(data)
plot.show()
def scatter(Attribute1 , Attribute2):
pkl_file = open('heart.pkl', 'rb')
dataset= pickle.load(pkl_file)
pkl_file.close()
data1=[]
data2=[]
Attributes_names=['Age','Sex','Chest','Resting_blood_pressure','Serum_cholestoral','Fasting_blood_sugar','Resting_electrocardiographic_results','Maximum_heart_rate_achieved','Exercise_induced_angina','Oldpeak','Slope','Number_of_major_vessels','Thal']
for i in range(0,len(Attributes_names)):
if Attributes_names[i]==Attribute1:
for instance in dataset:
data1.append(float(instance[i]))
if Attributes_names[i]==Attribute2:
for instance in dataset:
data2.append(float(instance[i]))
fig1, ax1 = plot.subplots()
ax1.set_title(Attribute1+" "+Attribute2)
ax1.scatter(data1,data2, facecolor='blue', alpha=1)
plot.show()
return np.corrcoef(data1, data2)
print(scatter("Age" , "Resting_blood_pressure"))DM Techniques
PART 1 : FREQUENTE ITEMS
Data discretization : (Equal intervals)
- Choose the number of partition
- Calculate the diffirence between Min and Max
- Split the interval on the number of partition and assign your data to there partitions
For more informations visit : http://magrit.cnrs.fr/docs/discretisation_fr.html
PART 1 : FREQUENTE ITEMS
Apriori
Two parametres
Min_support
Min confidence
Eclat
PART 1 : FREQUENTE ITEMS
Apriori
Exemple :
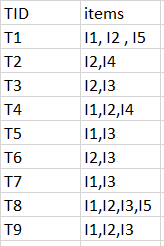
minimum support count is 2
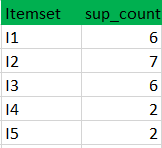

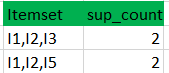
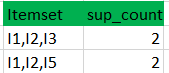
k=1
C1
L1
Data set
k=2
C2
L2
C3
L3
PART 1 : FREQUENTE ITEMS
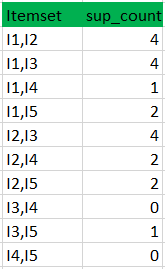
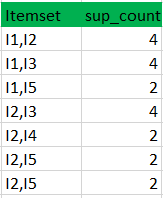
Apriori
Exemple :
minimum confidence is 50%
[I1^I2]=>[I3] //confidence = sup(I1^I2^I3)/sup(I1^I2) = 2/4*100=50% taken
[I1^I3]=>[I2] //confidence = sup(I1^I2^I3)/sup(I1^I3) = 2/4*100=50% taken
[I2^I3]=>[I1] //confidence = sup(I1^I2^I3)/sup(I2^I3) = 2/4*100=50% taken
[I1]=>[I2^I3] //confidence = sup(I1^I2^I3)/sup(I1) = 2/6*100=33% ignored
[I2]=>[I1^I3] //confidence = sup(I1^I2^I3)/sup(I2) = 2/7*100=28% ignored
[I3]=>[I1^I2] //confidence = sup(I1^I2^I3)/sup(I3) = 2/6*100=33% ignored
Confidence(A->B) = Support_count(A∪B) / Support_count(A)
PART 1 : FREQUENTE ITEMS
Eclat :
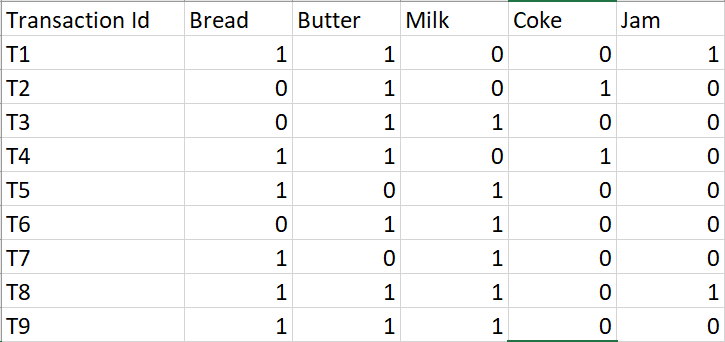
k = 1, minimum support = 2
Dataset
k = 2, minimum support = 2
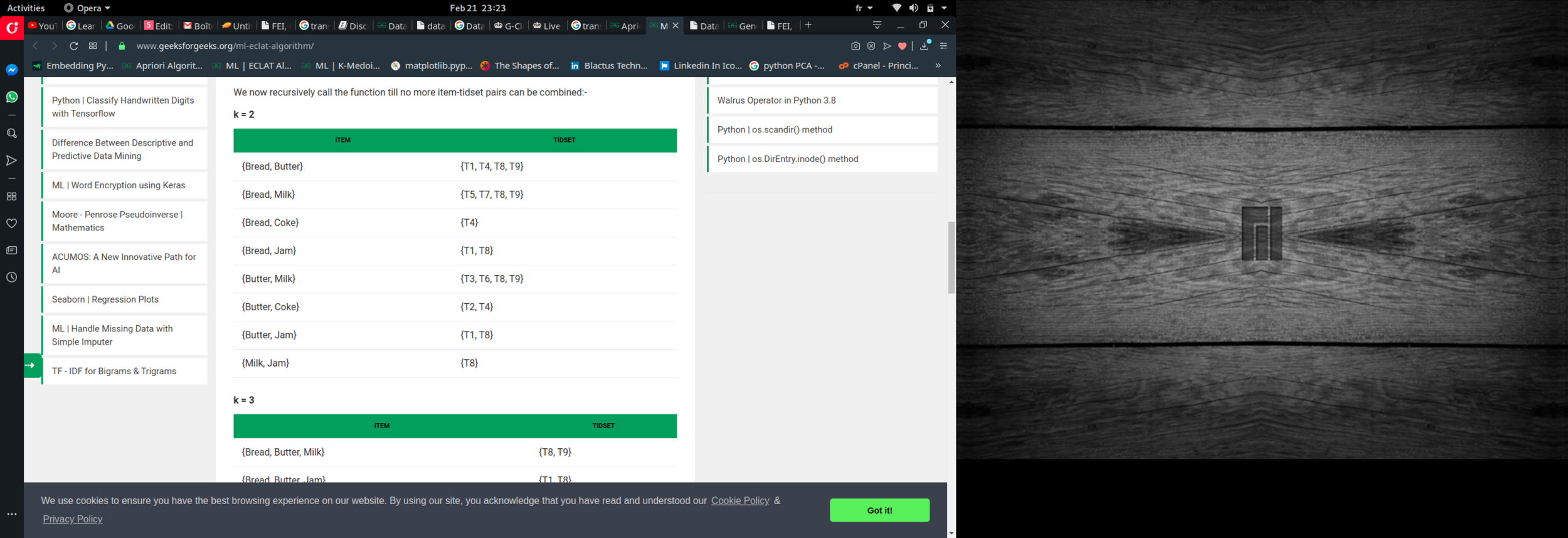
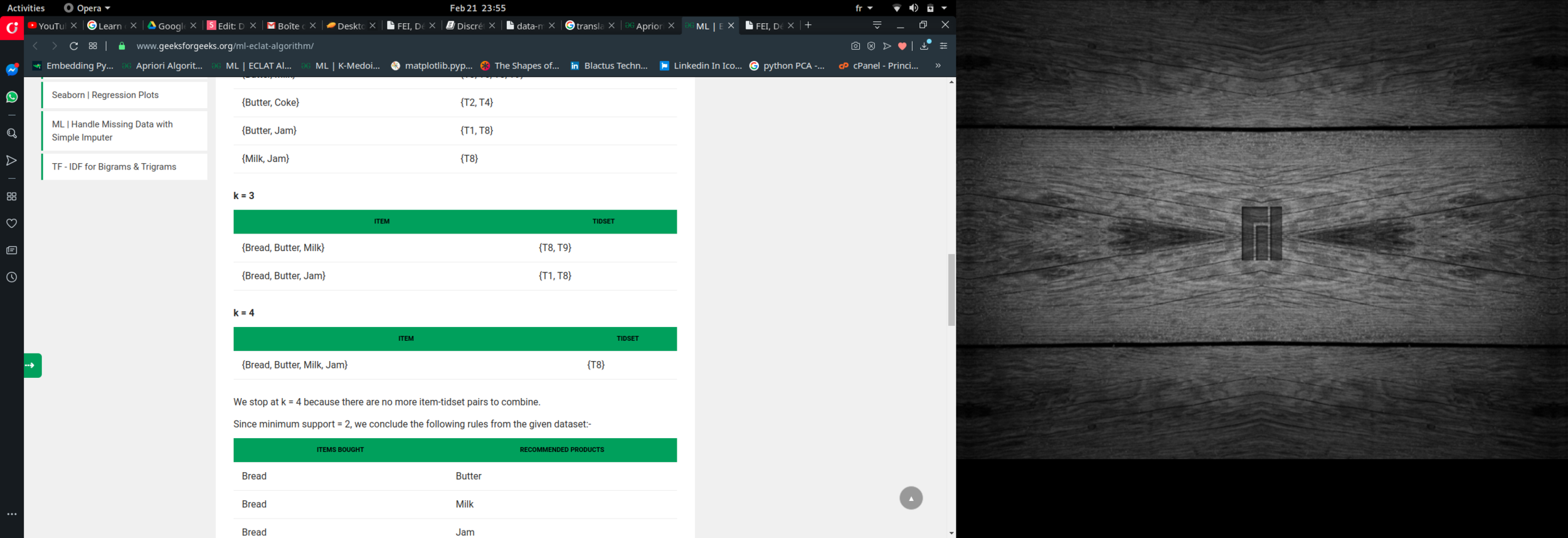

k = 3, minimum support = 2
k = 4, minimum support = 2
PART 2 : Unsuporvised classification
Data standarization : (min max methode)
new_value = old_value - min / max - min
Distance between data
d = sqrt ( (x_2 - x_1)² + (y_2 - y_1)² )
PART 2 : Unsuporvised classification
K-Means : k : clusters number
k=clusters_number
medeoids = [ k rundom data from the dataset]
for d in dataset:
calculate the distance between d and each medoid
assign d to the nearest medoid cluster
for c in clusters:
calculate the new medoid
boolean = false
while( not boolean):
recall k-mean with the new medoids
if clusters dont change:
boolean = true
PART 2 : Unsuporvised classification
Other algorithms :
- K-medoid
- Clara
- Clarans
Clusters visualization :
- PCA algorithms
Algorithms of optimisation :
- Genetic algorithm
import pickle
pkl_file = open('heart.pkl', 'rb')
dataset= pickle.load(pkl_file)
pkl_file.close()
age=[]
resting_blood_pressure=[]
serum_cholestoral=[]
maximum_heart_rate_achieved=[]
for i in range (0,len(dataset)):
age.append(float(dataset[i][0]))
resting_blood_pressure.append(float(dataset[i][3]))
serum_cholestoral.append(float(dataset[i][4]))
maximum_heart_rate_achieved.append(float(dataset[i][7]))
#age descritization
min_age = min(age)
max_age = max(age)
ecart_age = (max_age - min_age ) / 4
for i in range(0,len(dataset)):
if age[i]< min_age+ecart_age:
age[i]="age_1"
else:
if ( age[i] < min_age+2*ecart_age):
age[i]="age_2"
else:
if( age[i] < min_age+3*ecart_age):
age[i]="age_3"
else:
age[i]="age_4"
#resting_blood_pressure normalisation
min_resting_blood_pressure= min(resting_blood_pressure)
max_resting_blood_pressure= max(resting_blood_pressure)
ecart_resting_blood_pressure = (max_resting_blood_pressure - min_resting_blood_pressure ) / 4
for i in range(0,len(dataset)):
if resting_blood_pressure[i]< min_resting_blood_pressure+ecart_resting_blood_pressure:
resting_blood_pressure[i]="rbp_1"
else:
if ( resting_blood_pressure[i] < min_resting_blood_pressure+2*ecart_resting_blood_pressure):
resting_blood_pressure[i]="rbp_2"
else:
if( resting_blood_pressure[i] < min_resting_blood_pressure+3*ecart_resting_blood_pressure):
resting_blood_pressure[i]="rbp_3"
else:
resting_blood_pressure[i]="rbp_4"
#serum_cholestoral normalisation
min_serum_cholestoral = min(serum_cholestoral)
max_serum_cholestoral = max(serum_cholestoral)
ecart_serum_cholestoral = (max_serum_cholestoral - min_serum_cholestoral) / 4
for i in range(0,len(dataset)):
if serum_cholestoral[i]< min_serum_cholestoral+ecart_serum_cholestoral:
serum_cholestoral[i]="sc_1"
else:
if ( serum_cholestoral[i] < min_serum_cholestoral+2*ecart_serum_cholestoral):
serum_cholestoral[i]="sc_2"
else:
if( serum_cholestoral[i] < min_serum_cholestoral+3*ecart_serum_cholestoral):
serum_cholestoral[i]="sc_3"
else:
serum_cholestoral[i]="sc_4"
#maximum_heart_rate_achieved normalisation
min_maximum_heart_rate_achieved = min(maximum_heart_rate_achieved)
max_maximum_heart_rate_achieved = max(maximum_heart_rate_achieved)
ecart_maximum_heart_rate_achieved = (max_maximum_heart_rate_achieved - min_maximum_heart_rate_achieved) / 4
for i in range(0,len(dataset)):
if maximum_heart_rate_achieved[i]< min_maximum_heart_rate_achieved+ecart_maximum_heart_rate_achieved:
maximum_heart_rate_achieved[i]="max_hra_1"
else:
if ( maximum_heart_rate_achieved[i] < min_maximum_heart_rate_achieved+2*ecart_maximum_heart_rate_achieved):
maximum_heart_rate_achieved[i]="max_hra_2"
else:
if( maximum_heart_rate_achieved[i] < min_maximum_heart_rate_achieved+3*ecart_maximum_heart_rate_achieved):
maximum_heart_rate_achieved[i]="max_hra_3"
else:
maximum_heart_rate_achieved[i]="max_hra_4"
desc_data=[]
for i in range(0,len(age)):
desc_data.append([age[i],resting_blood_pressure[i],serum_cholestoral[i],maximum_heart_rate_achieved[i]])
for d in desc_data:
print(d)
output = open('desc_data.pkl', 'wb')
pickle.dump(desc_data, output)
output.close()THE END
Thanks for your attention
Contact :
email : soheib.boudali@blactus.tech linkedin : Soheib BOUDALI facebook : Cédric so instagram : cedric_s
https://blactus.tech
DM
By Soheib Boudali
DM
- 379



