Bioinformatics Team
MRC Clinical Sciences Centre
Thomas Carroll
Head Of Bioinformatics
The Bioinformatics Team.
- Tom Carroll
- Gopuraja Dharmalingam
- Sanjay Khadayate
- Yi-Fang Wang
- Yi-Wah Chan
- Marion Dore
- TBD
Websites
Where to find the team.
- ICTEM
- 2nd floor, MRC.
- Central aisle,
- Behind the printers.
Role
- Analysis
- Experimental design.
- Bioinformatics Infrastructure.
- Training.
- Bioinformatics Seminar Series
Experimental Design
“To consult the statistician after an experiment is finished is often merely to ask him to conduct a post mortem examination. He can perhaps say what the experiment died of.”
Fisher RA, 1938
- Work closely with Genomics Team to help with design questions
- Replicate number.
- Sequencing depth.
- Sequencing strategy.
Nice example experimental design
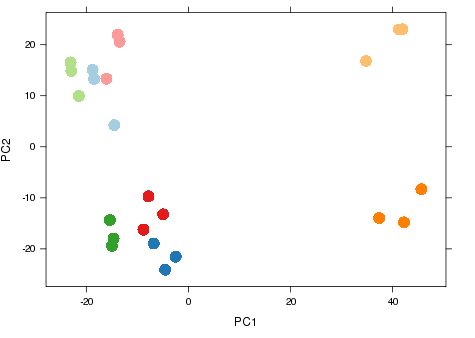
- RNA-seq experiment (2014)
- Graph shows major sources of variation.
- Samples from same groups close together.
- Samples from different experimental conditions separate.
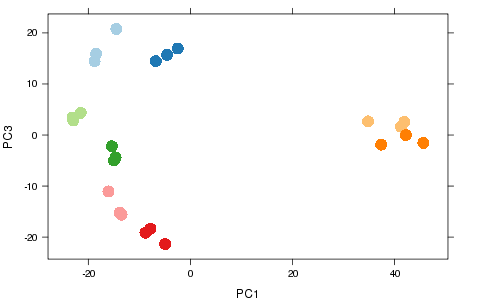
Nice example of experimental design
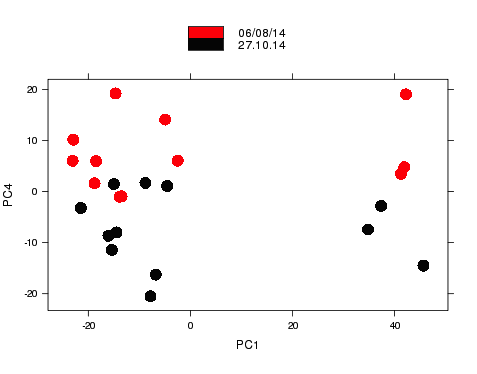
- Smaller sources of variance relating to other metadata.
- Samples group according to the day that RNA was extracted on.
- Known effects can be removed from analysis.
Analysis
- Initial data processing and QC.
- Advice and support as needed.
- Support throughout project.
Increased demand for long term support.
Authorships in 28 publications since 2014.
Analysis support
- Increased use of high throughput techniques in projects.
- Greater use for bioinformatics in projects.
- Analysis across project lifetime or individual elements.
- Requires reproducible research.
Reproducible research
- Reproducible results from computational methods should be straight forward.
- Common problems.
- Version and software changes.
- Lack of analysis documentation.
rMarkdown
- rMarkdown converted R code to dynamic reports.
- Code, results and versions are reported within the same page.
- HTML allows for inclusion of dynamic elements.
A do it yourself guide
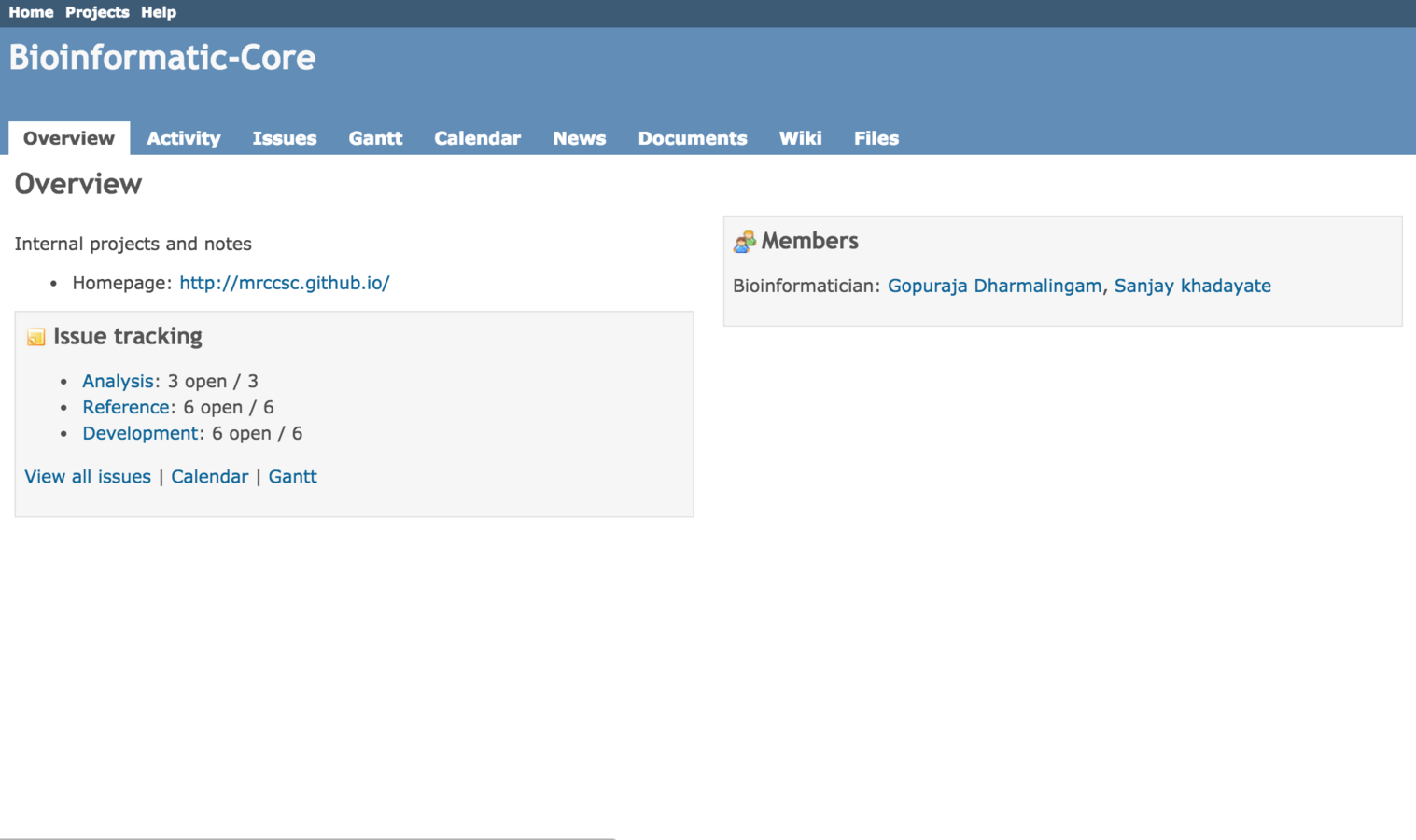
Project tracking
- Use Redmine software.
- Multiple user interface to record project information.
- Repository to version control scripts (SVN).
- Wiki for internal documentation.
Infrastructure
- Analysis pipelines.
- Data delivery.
- Software development.
Basecalling/Demultiplexing, ChIP-seq and RNA-seq
pipelines.
- Common analysis steps can be automated.
- Optimised for local resources.
- Reproducible and comparable.
- Basecalling, ChIP-seq and RNA-seq pipeline to automate sequence acquisition, demultiplexing, alignment and quality control.
- Freely available for use or customisation on github
RNA-seq and others in the pipeline
- Internal RNA-seq pipeline
- Written in R.
- Easily installed, maintained.
- Allows Core to move between systems easily.
- Released soon.
- Genomics pipeline.
- R based.
- Automate basecalling and sequence QC capture.
- Development version on github site.
- ChIP-seq R pipeline.
- Basecalling to ChIP/RNA-seq QC.
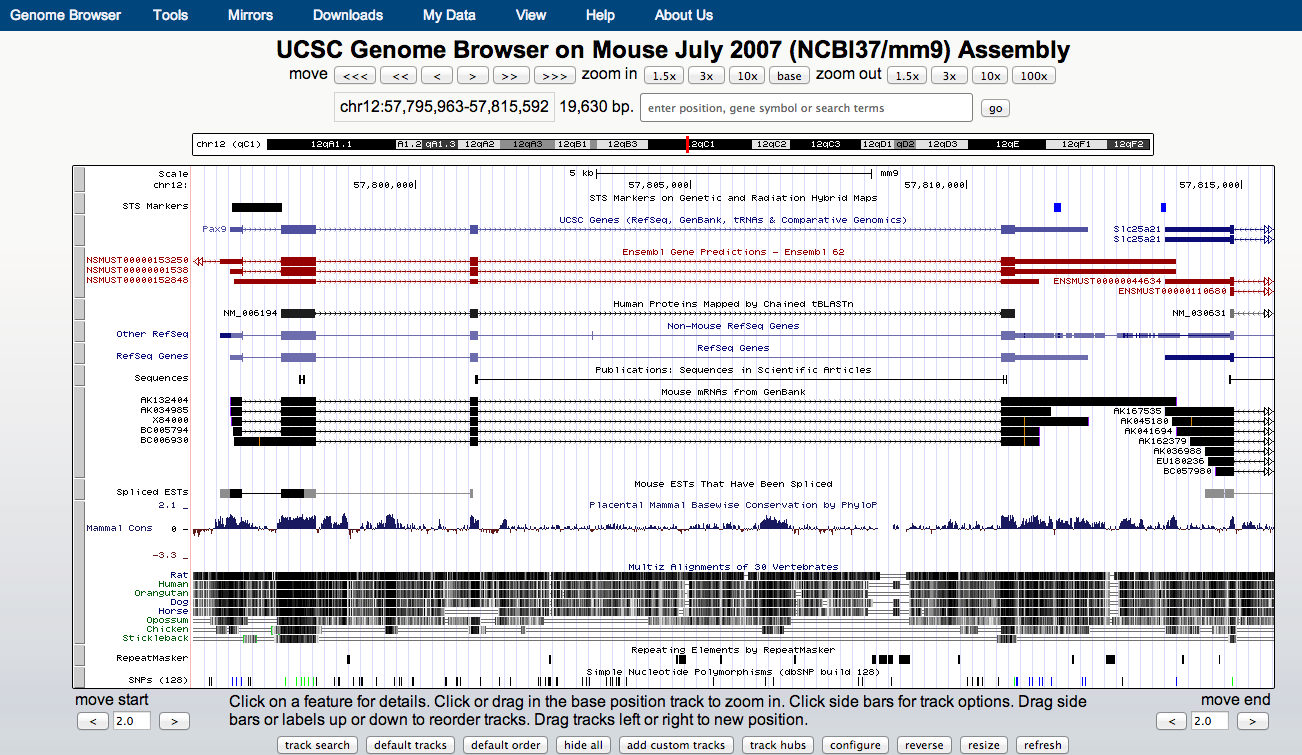
UCSC genome browser
- UCSC allows for visualisation of a range of genomics data types.
- Public instances can be very slow.
- CSC public instance maintained by Bioinformatics team.
-
web: http://ucsc
FTP: ftp://ucsc
Software
- Develop and maintain software relevant to our work.
- R packages and javascript toolsets.
-
Release software to public (peer-reviewed) repositories.
- Collaborative feedback.
- Automated build reports and checking.
ChIPQC
- Lack of suitable R/Bioconductor quality control tools for ChIP-seq.
- Require methods to assess quality across high volumes of samples
- ChIPQC developed and tested on 500 public datasets.
- IGV is an popular alternate to UCSC.
- Allows for inclusion of per sample metadata and complex sample display types.
- Tracktables creates standalone and rMarkdown compliant tables.
Tracktables
- Visualising genomics data over regions of the genome.
- Allows for rapid generation of profiles and subsetting by IDs or other regions.
- Arithmetic operations between and within profiles allows for rapid, iterative investigation of hypotheses.
Soggi
- Peak calling in R is convenient.
- Many peak callers in R have unwieldy input and far from optimised.
- triform contains a reliable peak calling algorithm in need of optimisation for speed and long marks.
- MRC CSC took over maintenance of triform in 2015
triform
Training
-
Aim to develop courses to meet MRC Clinical Sciences requirements.
- R
- Python
- High throughput sequencing analysis.
CSC Bioinformatics Course
- Current and upcoming Bioinformatics training material can be found at our site
http://mrccsc.github.io/training.html
Training Collaborations
Develop and share courses between other Bioinformatics teams.
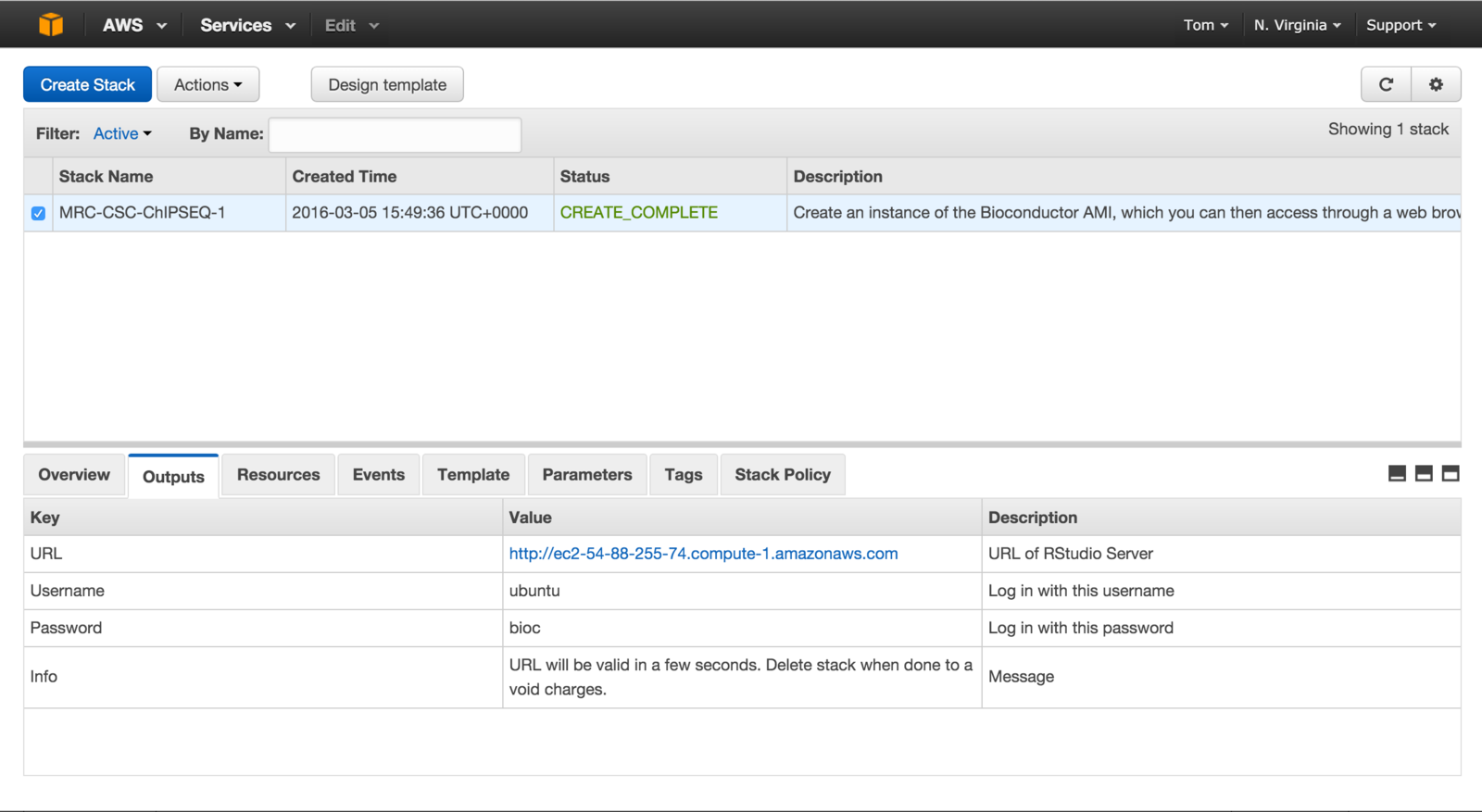
Training on the cloud.
- Awarded grant from Amazon Web Services.
- Use virtual linux servers to host R and RStudio pre-loaded with course material.
- Allow for larger, real world analysis tasks during training.
- No need for dedicated classroom - train from anywhere.
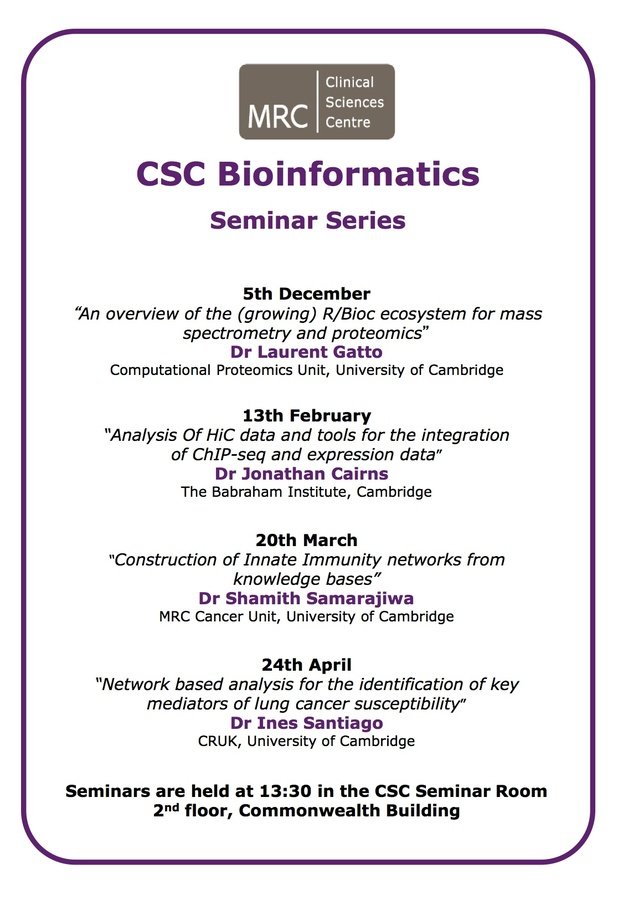
Bioinformatics Seminar Series
-
Features external and internal speakers.
-
Discuss methodology behind bioinformatics analyses.
Bioinformatics Seminar Series
Laurent Gatto
Johnathan Cairns
Shamith Samarajiwa
Ines de Santiago
5th, December
13th, February
20th, March
24th, April
Head of Computational Proteomics Unit, Cambridge
Postdoc, Peter Frasier lab, Babraham Institute
Prinicipal Investigator, MRC Cancer Unit
Postdoc, Markowetz lab, CRUK.
Contacts and thanks
Bioinformatics Team
Tom - thomas.carroll@imperial.ac.uk
Gopu - gopuraja.dharmalingam@imperial.ac.uk
Sanjay - sanjay.khadayate@imperial.ac.uk
Yi-Fang - yifang.wang@imperial.ac.uk
Marion - marion.dore@imperial.ac.uk
Yi-Wah Chan - y.chan@imperial.ac.uk
Bioinforma
By tom carroll
Bioinforma
- 674



