Short Intro to
shRNA Single-Cell
RNA-seq
Sample Information
- 96 Samples
- Single Lane ?- 274.44 M reads

Processing Steps
- RNA-seq
- IGenomes UCSC mm9
- Tophat 2 Unstranded with ERCC spikes
- FeatureCounts only paired ---Rsubread
- DESEQ2 - Standard analysis
- using RUVseq to get spike in adjustments.
- rlog transform for PCA.
Processing Steps
shRNA-seq
- pRLL_shRNA_Rep_library in csv
- Full shRNA FASTA contigs built in R
- Duplicate names removed.
- Indexed and aligned using bwa.
- Properly paired and unique mapping hits retained
- Counts for shRNAs in samples established from mapped BAMS.
shRNA
- Alignments to shRNA inspected in IGV.
- shRNA alignments realigned to genome.
- Cross contamination of reads in shRNA to target gene and reverse.
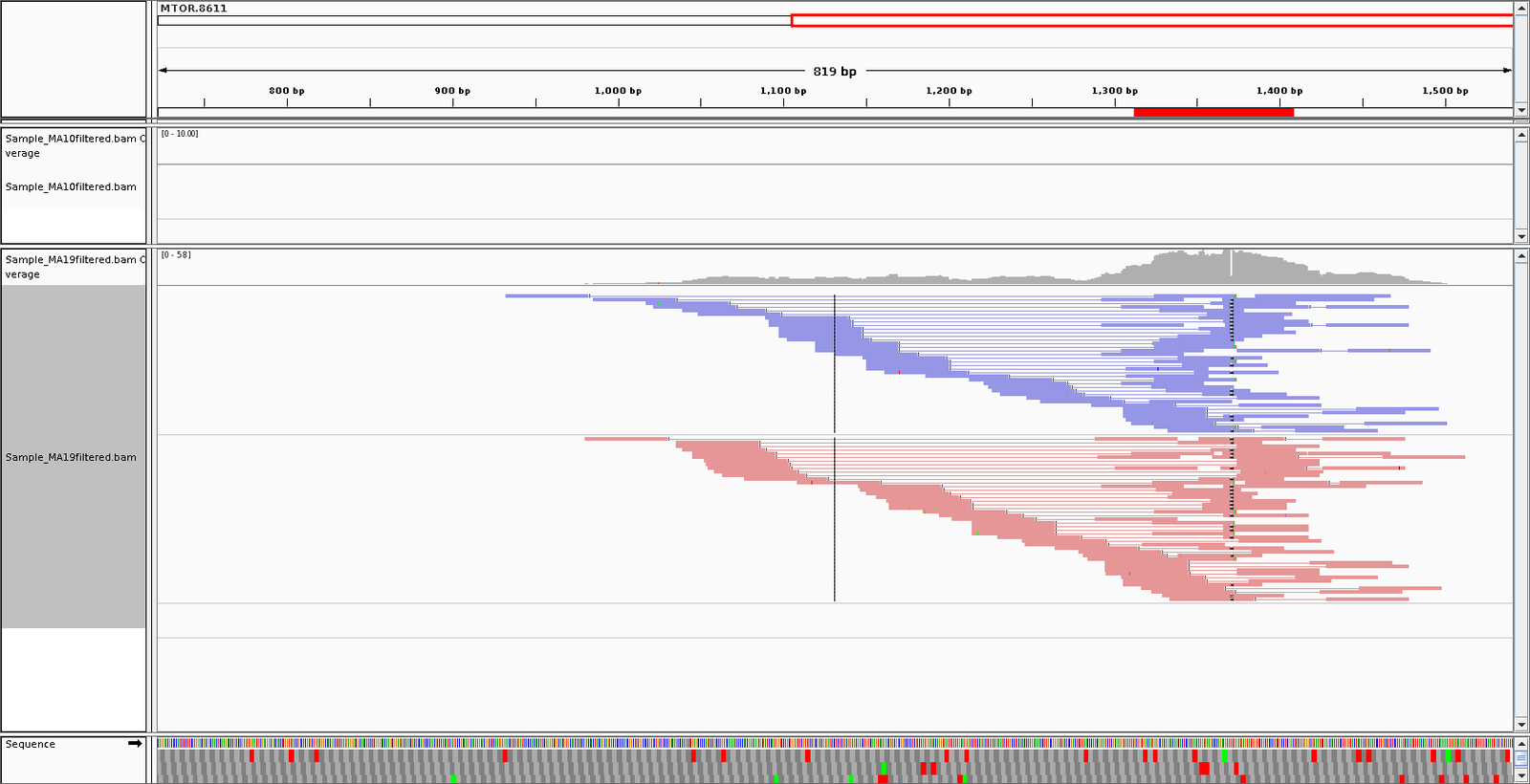
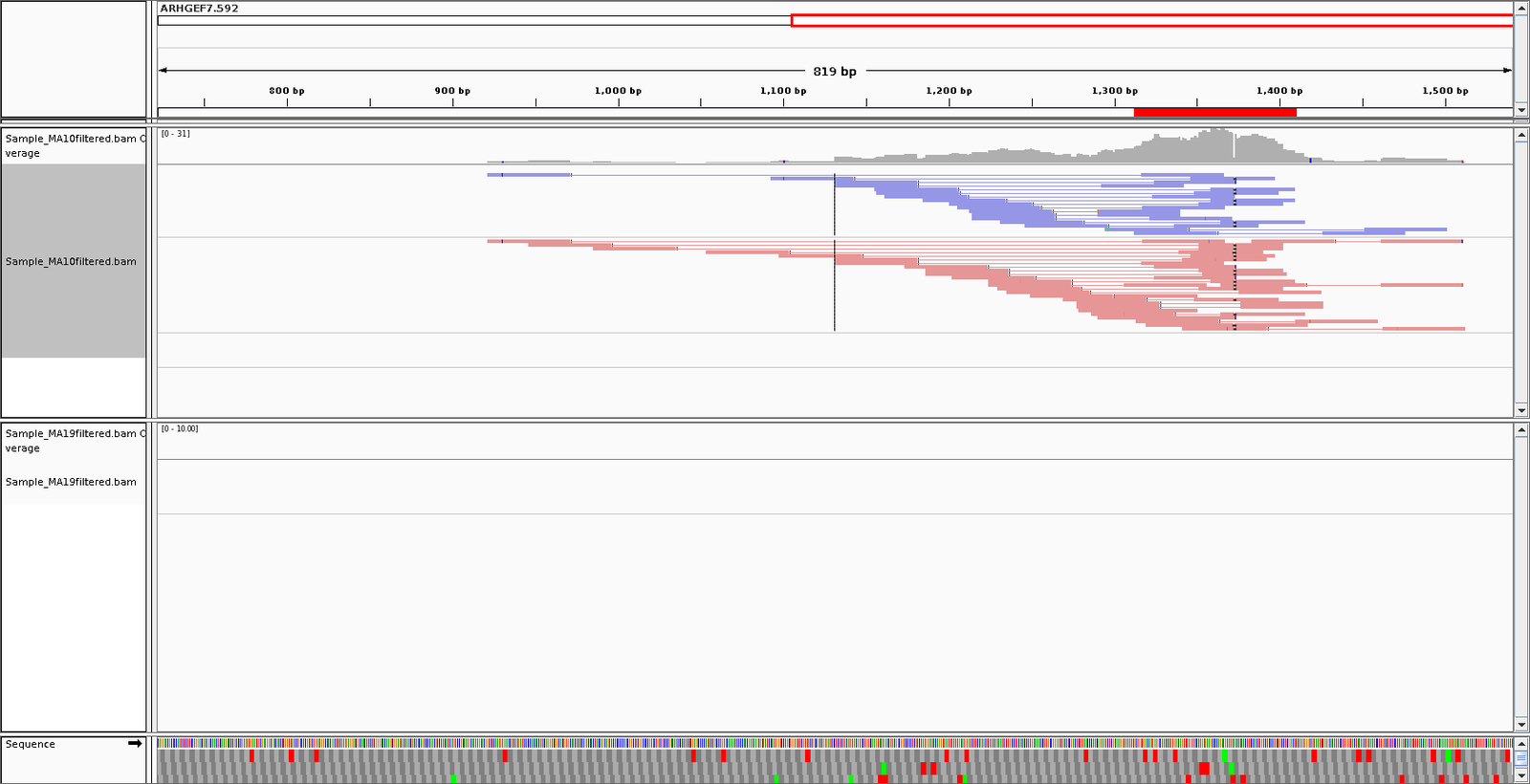
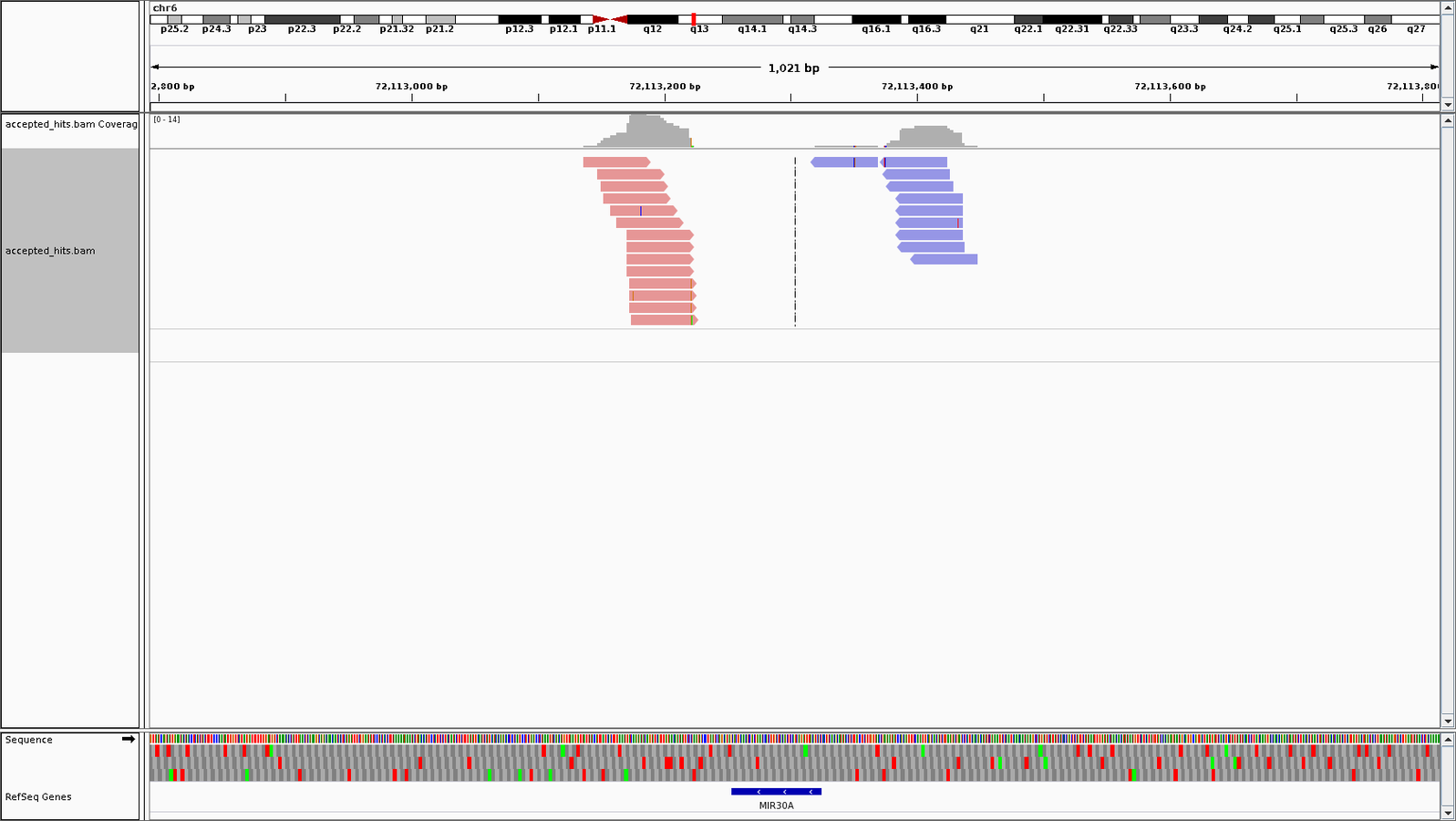
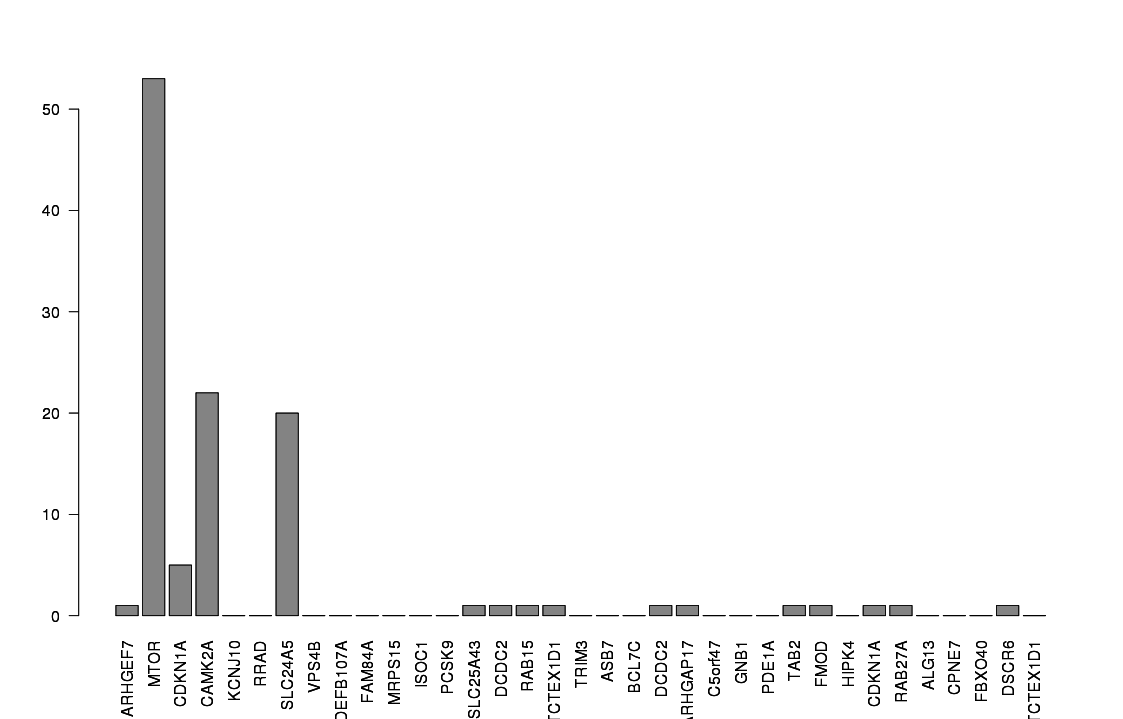
Distribution of shRNAs
RNA-seq
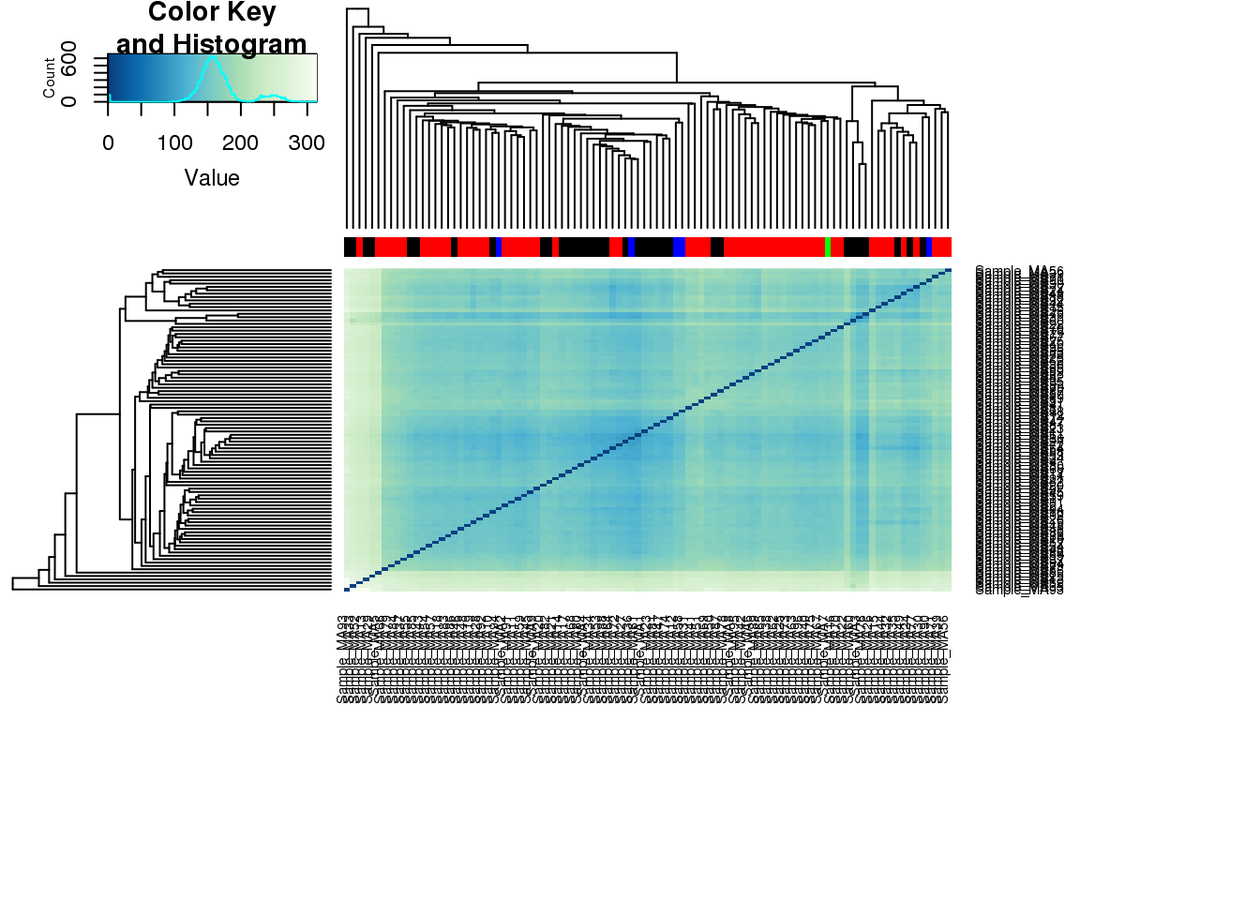
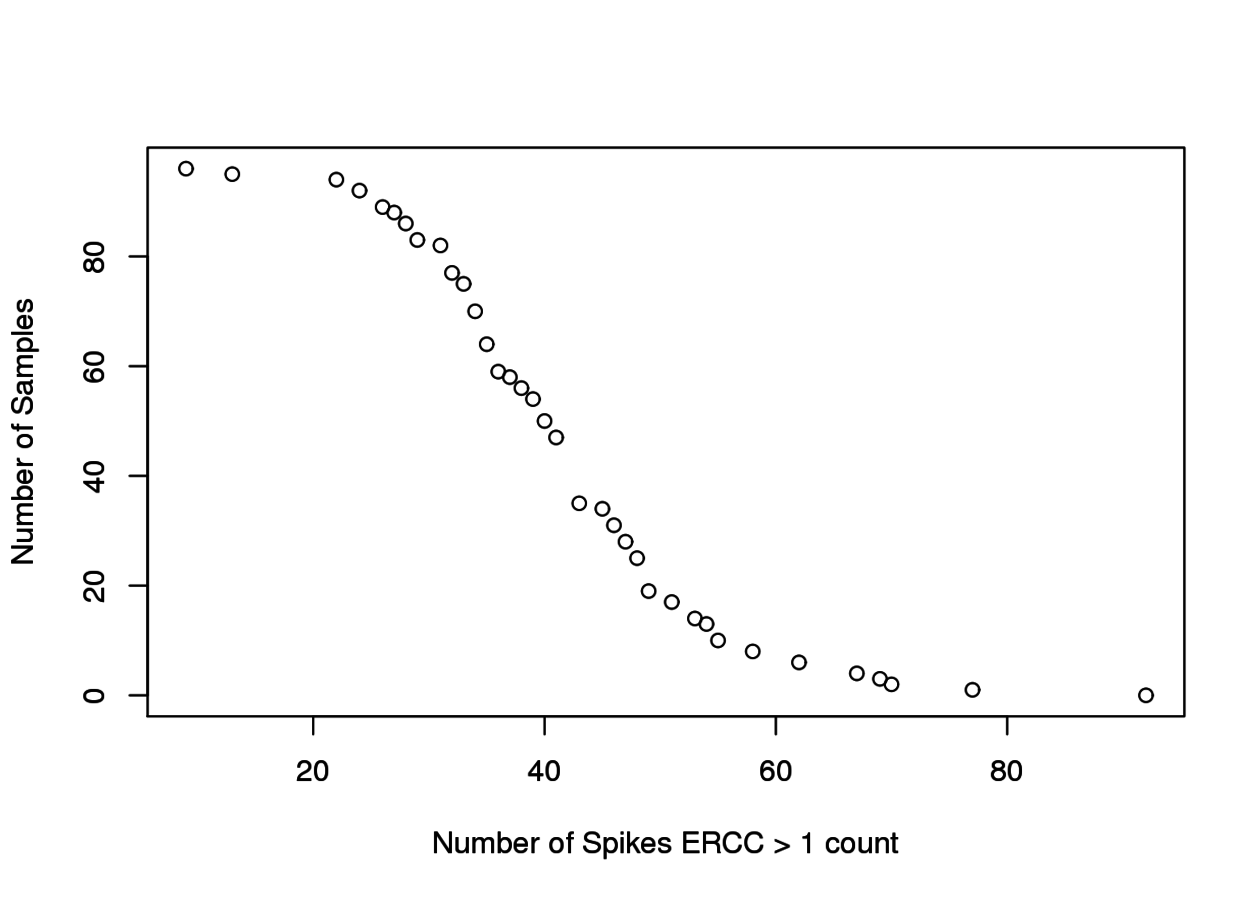
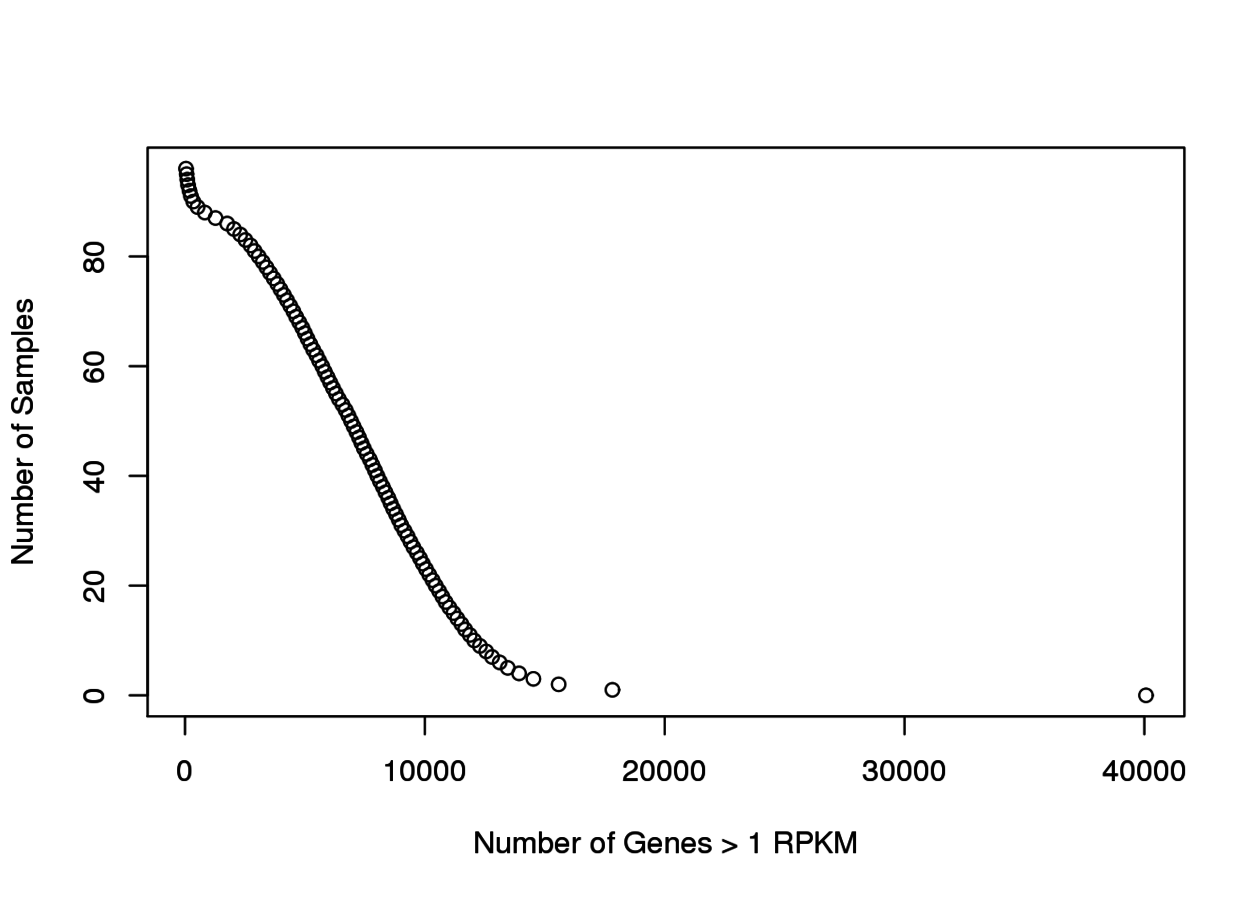
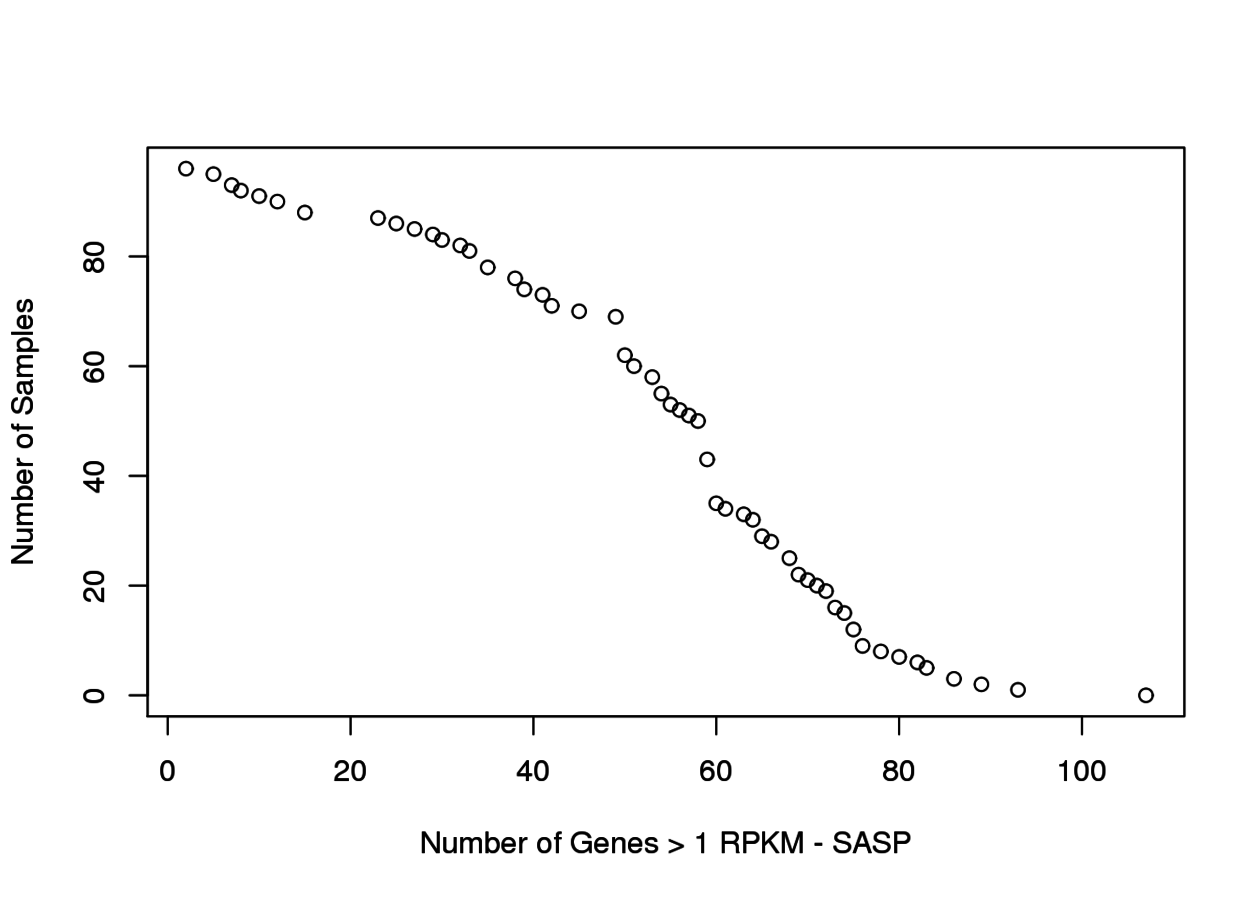
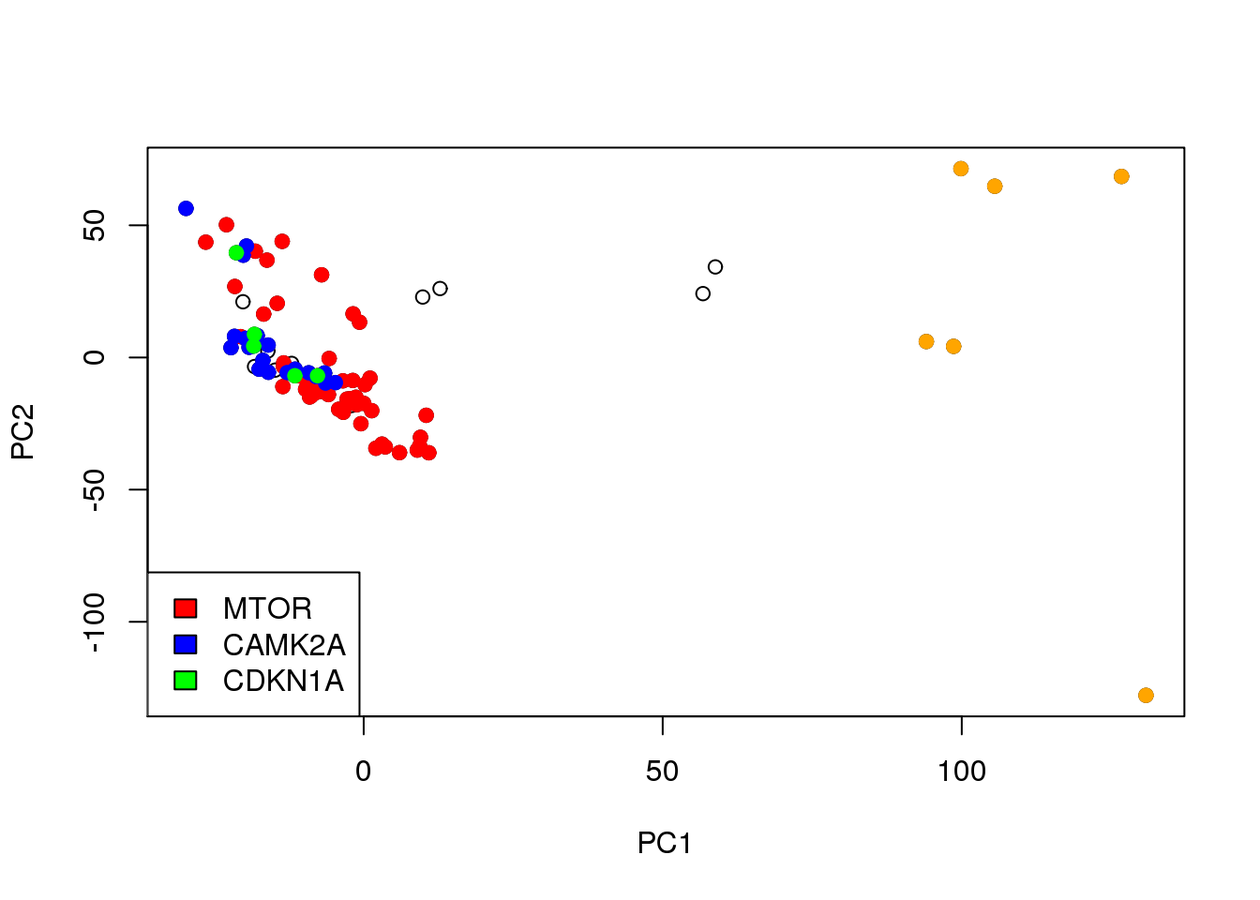

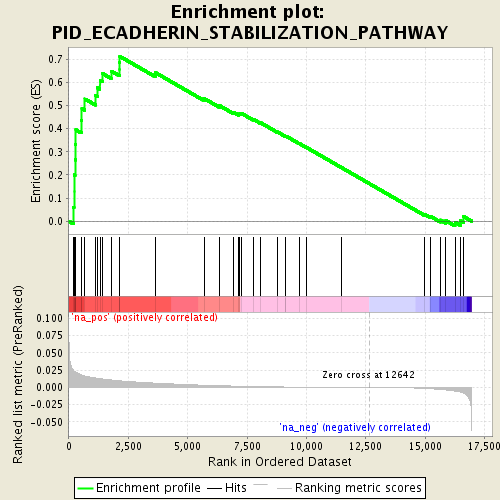
Associated to MTOR -ve PC2 loading
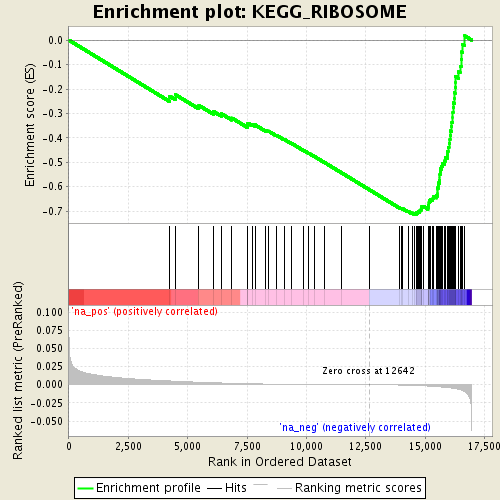
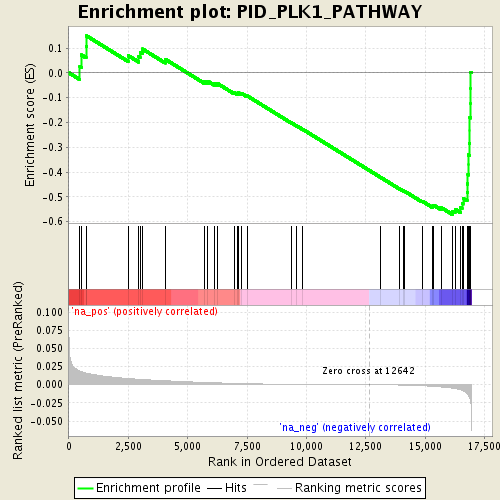
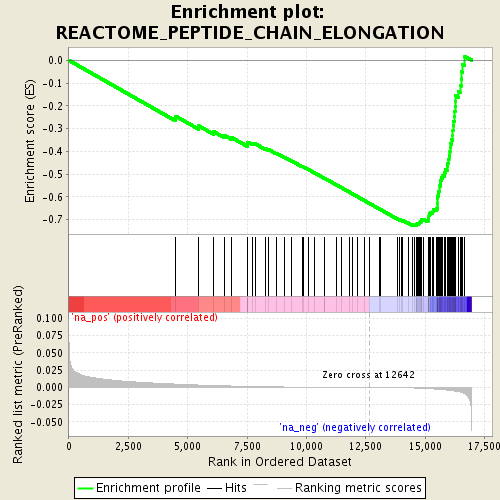
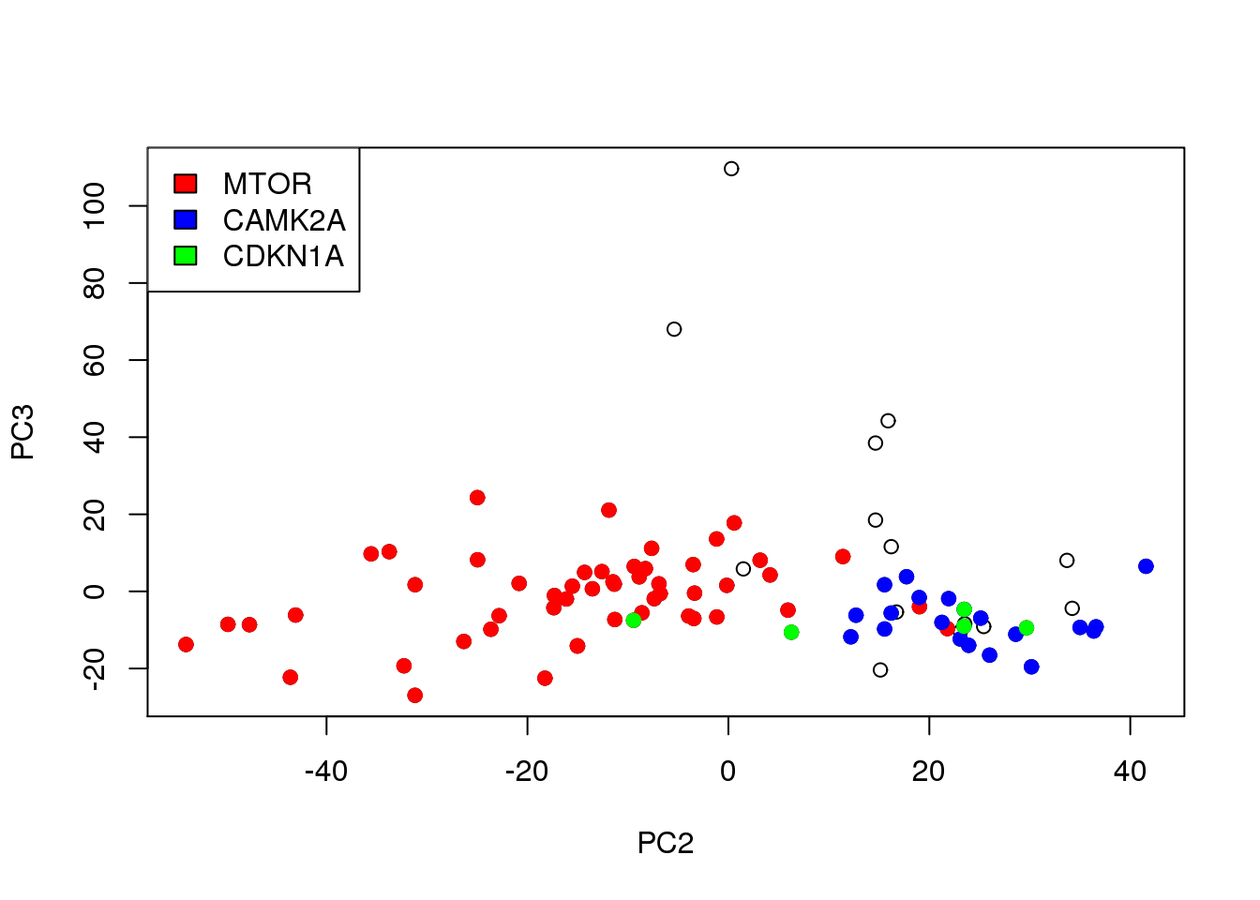
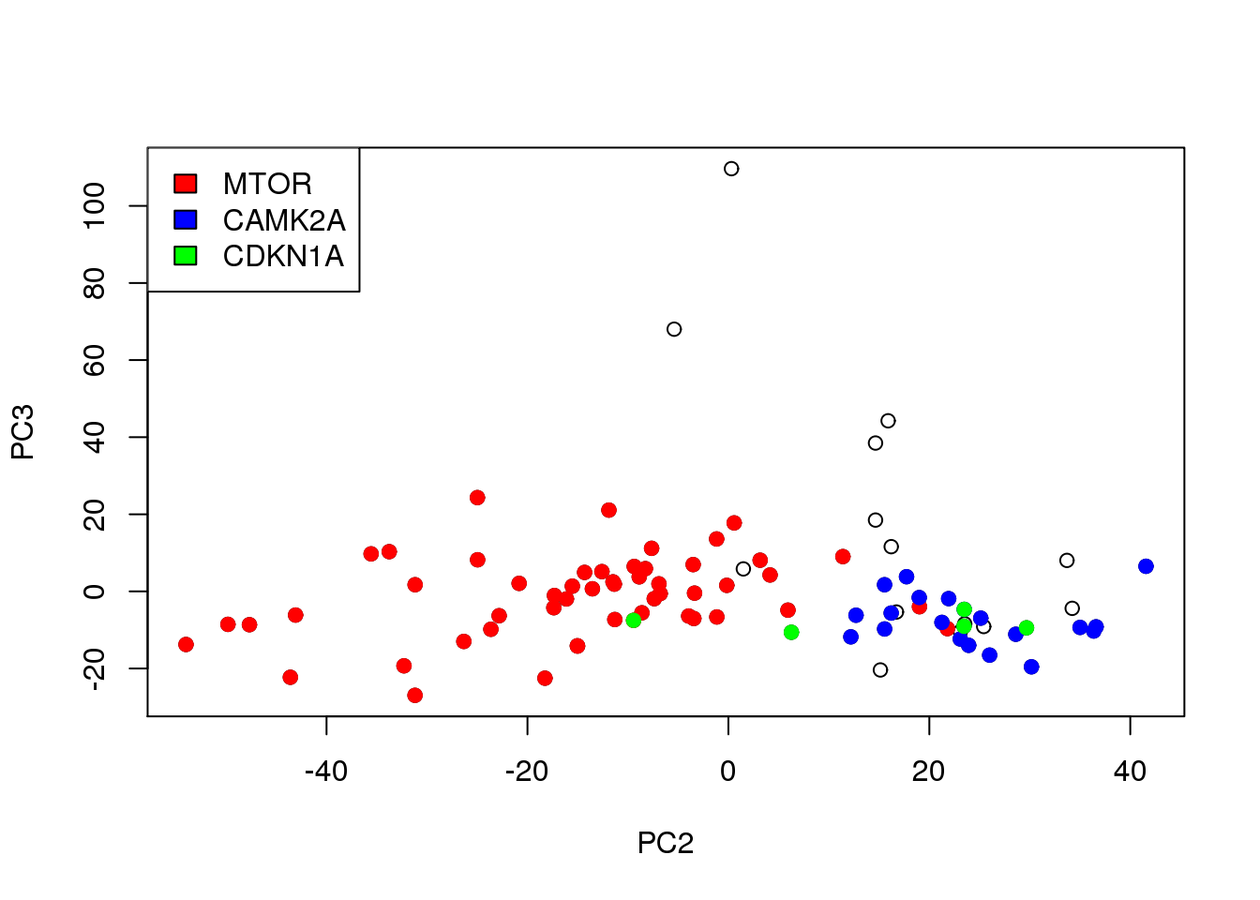
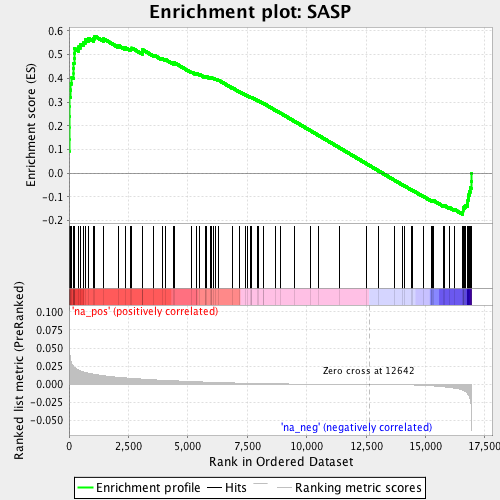
Associated to Cdkn1a/Camk2a

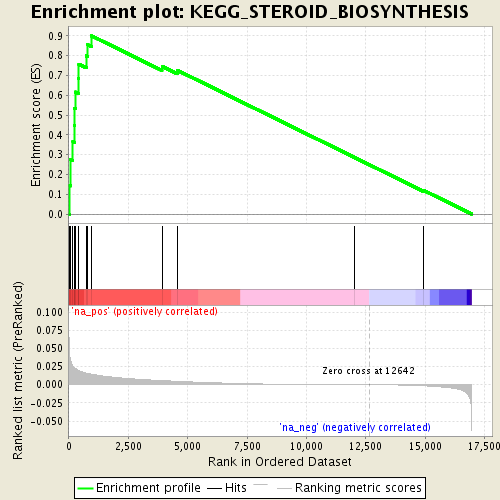
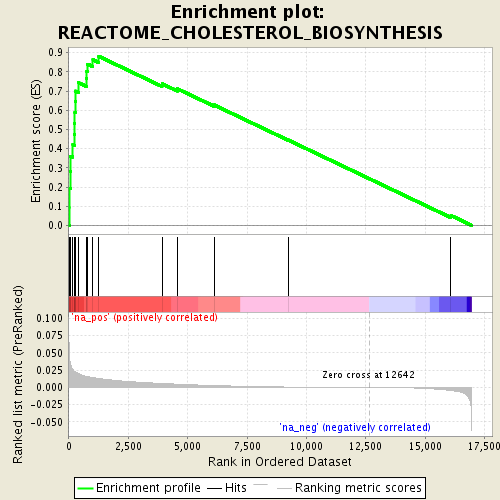
singleCell
By tom carroll
singleCell
- 598



