Parallelization of a BioInformatics
program in Python
Participant:
-
Zaika Vladyslav
Supervisors:
-
Denis Pallez
-
Claude Pasquier
miRAI beat cancer
microRNA - nucleotides, that regulate thousands of human genes
miRNA desregulations related to development of various diseases (CANCER)
miRAI predicts associations between miRNA and diseases.
Problem
miRAI uses many parameters (37) to perform predictions
all combination of parameters represents:
Computation of one case takes ~ 4min - 4hours
Solution
Distribute miRAI computations on cluster
Genetic algorithms to accelarate computations
Assigned tasks
1. Select parallel evolutionary python fw
2. Perform computations on one node
3. Configure cluster nodes
4. Distribute computation to cluster
5. Compare frameworks
6. Propose improvements
Inspyred
FW for evolutionary computations
Connected with PP module
Inspyred
Adapted for local networks
Quick bootstrap
Scheduler issue
on the nodes: node-1> ./ppserver.py -a node-2> ./ppserver.py -a
final_pop = ea.evolve(generator=generate, evaluator=inspyred.ec.evaluators.parallel_evaluation_pp,
pp_evaluator=evaluate,
pp_servers=("*",),
pp_dependencies=(my_squaring_function,),
pp_modules=("math",),
pop_size=8,
bounder=inspyred.ec.Bounder(-5.12, 5.12),
maximize=False,
max_evaluations=256,
num_inputs=3)
DEAP
FW created specially for parallel evaluation executions
Uses SCOOP for parallelism
Quebec, Laval university project
DEAP
Connects to any machine
No tuning on nodes
Hard to configure
Manual config of hosts
from scoop import futures
toolbox.register("map", futures.map)
python -m scoop --hostfile hosts program.py
hostname_or_ip 4
other_hostname
third_hostname 2
Cluster
Inspyred:
- fast start
- minimum code
- auto configuration
- single cluster
- scheduling
DEAP:
- no scripts on nodes
- scalability
- still active
- manual config
- hard to setup
PP vs SCOOP
PP
SCOOP
Scheduling
PP scheduler assign all the tasks in the begining
SCOOP scheduler wait until current task finishes
SCOOP + Inspyred
Dev scoop parallelism for inspyred
final_pop = my_ec.evolve(generator=generate,
evaluator=parallel_evaluator_scoop,
scoop_evaluator=evaluate,
pop_size=1,
maximize=True,
max_generations=5,
num_elites=_NumberOfElite,
seeds=None,
dimension_bits=_NumberOfBits
)
def evaluate(candidates, args):
fitness = []
for cs in candidates:
fit = miRAI.evaluate(params)
fitness.append(fit)
return fitness
def generate(random, args):
size = args.get('dimension_bits', 10)
return [random.choice((0,1)) for i in range(size)]
def parallel_evaluator_scoop(candidates, args):
evaluator = args['scoop_evaluator']
results = list(futures.map(evaluator, candidates, args))
return results
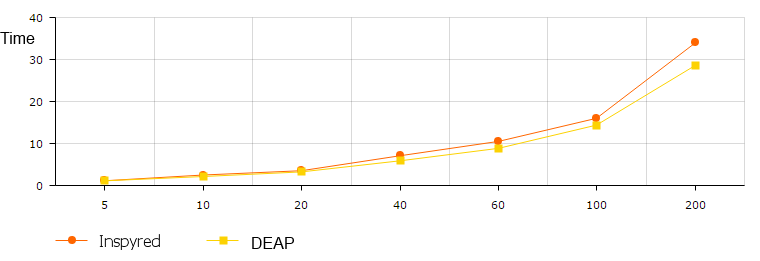
Benchmarking
- max execution time on node
- mean execution time on node
- check Amdahl's law:
Future steps
- Test SCOOP + Inspyred
- Put everything to cluster
- Calculate miRAI algorithm
- Proceed the results
OAR
OAR - task manager
jdoe@idpot:~$ oarsub -I -l /nodes=3/core=1
jdoe@idpot5:~$ cat $OAR_NODEFILE
idpot5.grenoble.grid5000.fr
idpot8.grenoble.grid5000.fr
idpot9.grenoble.grid5000.fr
#!/bin/bash
python3 insp_script.py
Conclusion
- Test Inpyred vs DEAP
- OAR configuration
- Detect weak sides
- SCOOP + Inspyred adhoc
- Benchmarks
Thank you
vlad@nowinfinity.com.au
Publications
Scientific paper: "Prediction of miRNA-disease associations with vector space model."
PFE presentation
By Vladyslav Zaika
PFE presentation
Parallelization of a Bioinformatics program in Python.
- 309



