LeafCutter

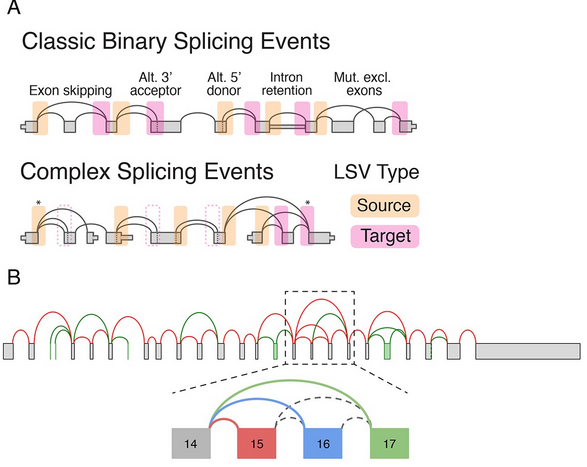
Several types of common alternatively splicing events are captured by the alternative excision of introns.



phastcons score: posterior probabilities that each site was generated by the conserved state





(C) Per nucleotide average conservation score (phastCons60 track) in regions proximal to single source (top) and single target (bottom) LSVs that were differentially spliced between any pair of tissues shown in Figure 4. The average is plotted for the subsets of complex (green) LSVs and binary (grey) LSVs as well as around a randomly selected set of constitutively spliced junctions (red, see Materials and methods for details).
tissue regulated splicing involves significantly higher conservation in the intron proximal to the variable exonic segments, a region known to include cis elements to which tissue specific splice factors bind
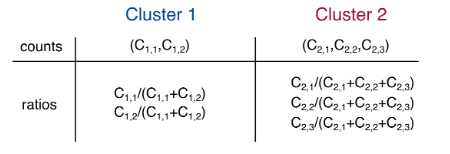
Percentage Splice Index (PSI): The ratio between inclusion reads and inclusion reads plus exclusion reads PSI = A + B / (A +B + C)







(a) LeafCutter uses split reads to uncover alternative choices of intron excision by finding introns that share splice sites. In this example, LeafCutter identifies two clusters of variably excised introns.
(b) LeafCutter workflow. First, short reads are mapped to the genome. When SNP data are available, WASP (van de Geijn et al., 2015) should be used to filter allele-specific reads that map with a bias. Next, LeafCutter extracts junction reads from .bam files, identifies alternatively excised intron clusters, and summarizes intron usage as counts or proportions. Lastly, LeafCutter identifies intron clusters with differentially excised introns between two user-defined groups using a Dirichlet-multinomial model or maps genetic variants associated with intron excision levels using a linear model.
(c) Using LeafCutter to discover novel introns, we find that for any given tissue, over 10% of alternatively excised introns are unannotated. Remarkably, 48.5% of testis alternatively excised introns are unannotated. Different colors denote the proportion of introns when one or more splice sites are unannotated “(ss absent)”, both splice sites are annotated but the intron is not part of any transcript “(ss present)”, or when the intron is annotated in some but not all databases.
(d) The unannotated splice sites of novel introns show moderate signature of sequence conservation as determined by vertebrate phastCons scores. Miss one: conservation of the unannotated splice site of an intron for which the cognate splice site is annotated. Miss both: conservation of splice sites of introns with both splice sites unannotated.


Hierarchical clustering on all 1,258 introns that had no missing values in any of the samples
(a) LeafCutter identifies tissue-regulated intron splicing events from GTEx organ samples. Heatmap of the intron excision ratios of the top 500 introns that were found to be differentially spliced between at least one tissue pair. Tissues include brain (Br), muscle (Ms), heart (Ht), blood (Bd), pancreas (Pc), esophagus (Eg), and testis (Ts).
(b) Tissue-dependent intron excision is conserved across mammals. Heatmap showing intron exclusion ratios of introns differentially spliced between pairs of tissues (Muscle vs Colon, Brain vs Liver, and heart vs Lung). Heatmap shows 100 random introns (97 for the heart vs lung comparison) that were predicted to be differentially excised in human with p-value < 10−10 (LR-test) and that had no more than 5 samples where the excision rate could not be determined due to low count numbers. Heatmap of all introns that pass our criteria can be found in Supplementary Figure S6.
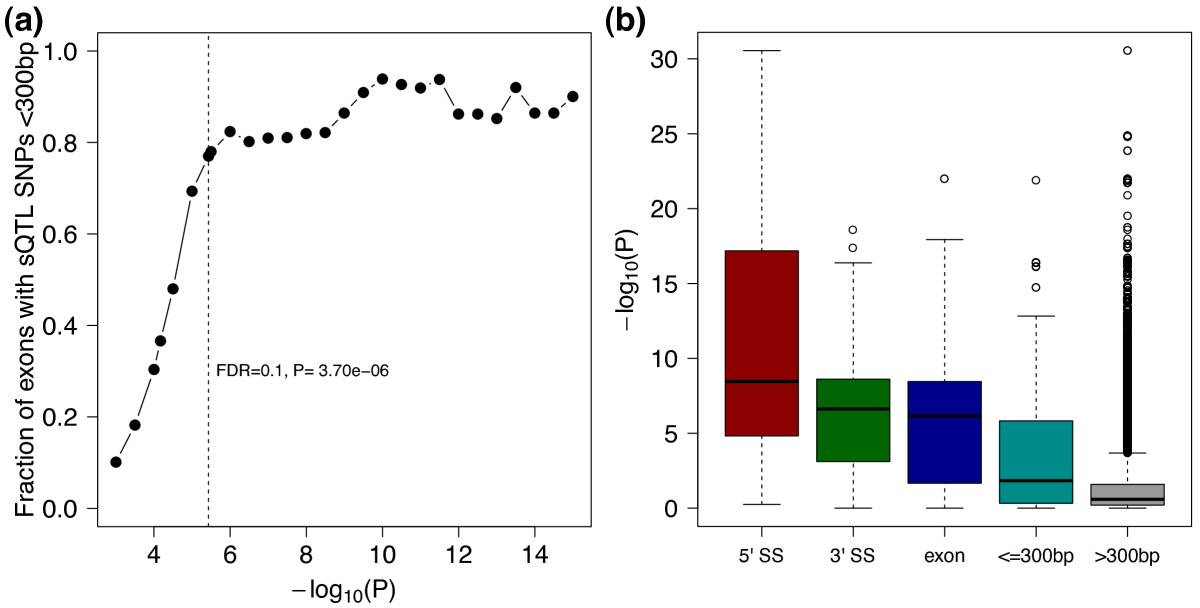
(c) QQ-plot showing genome-wide sQTL signal in LCLs (black), sQTL signal conditioned on exon eQTLs (purple) and conditioned on transcript ratio QTLs (dark purple) from (Lappalainen et al., 2013). Signal from permuted data in light grey shows that the test is well-calibrated.
(d) Positional distribution of sQTLs across LeafCutter-defined intron clusters. 1,421 of 4,543 sQTLs lie outside the boundaries (Supplementary Figure S7 for all sQTLs). (e) High proportion of shared sQTLs across four tissues from (Ardlie et al., 2015).
(f) Example of a SNP associated to the excision level of an intron in blood but not in other tissues.
(g) Enrichment of low p-value associations to multiple sclerosis and rheumatoid arthritis among LeafCutter sQTL and gEUVADIS eQTL SNPs. The numbers of top sQTLs and eQTLs that are tested in each GWAS are shown in parentheses.



Comparison between beta-binomial and Dirichlet-multinomial models for differential splicing analyses, performed on 10 male brain vs. heart samples from GTEx. Two approaches for combining perintron p-values into cluster level introns are compared: Bonferroni correction and Fisher’s combined test. Bonferroni is very conservative, as expected. Fisher’s combined test has considerably lower power than the multinomial approaches. However, only v2 of the Dirichlet-multinomial (which uses a per intron concentration/overdispersion parameter) is well calibrated under permutations.
LeafCutter
By acoutoal
LeafCutter
- 1,494



