Time series for DNA-DNA interactions
Sasha Galitsyna
Nikolai Bykov
SMTB 2021

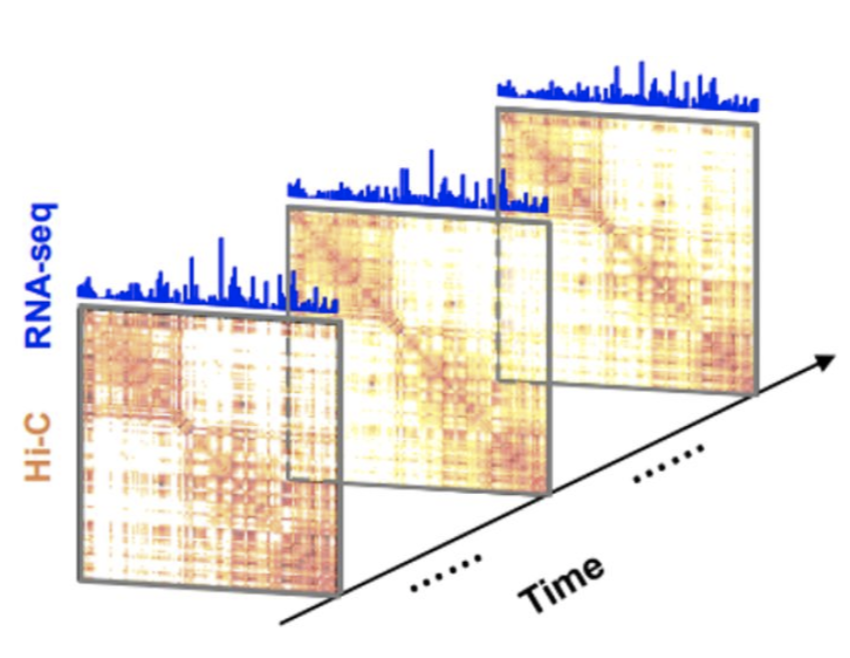
What do we need for successful rotation?
Google Colab:
https://colab.research.google.com/
Software:
https://github.com/encent/hichew/blob/master/examples/hichew_api_example.ipynb
Link to Google Drive with Hi-C data:
https://drive.google.com/drive/folders/16A0x0TMbxsO849kxFykJmZVXeE7Fq0K_?usp=sharing
These slides:
https://slides.com/agalicina/time-series-dna-graph
Chromatin spatial structure
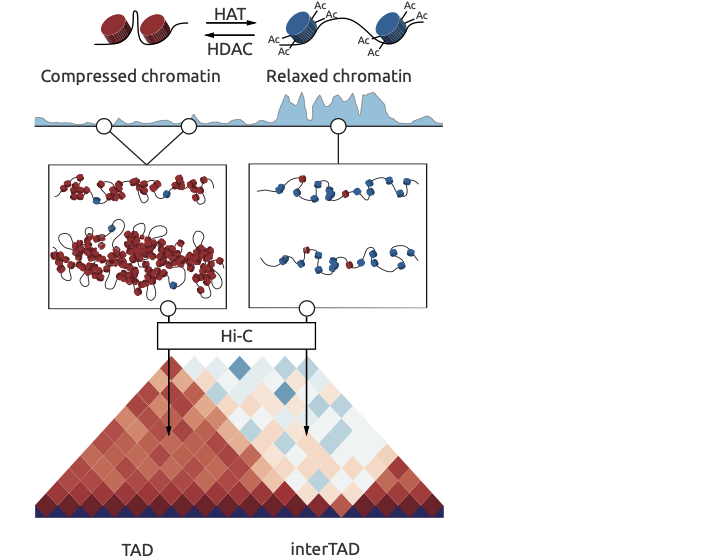
Ulianov et al. Genome Biology 2016

Methods to study chromatin structure
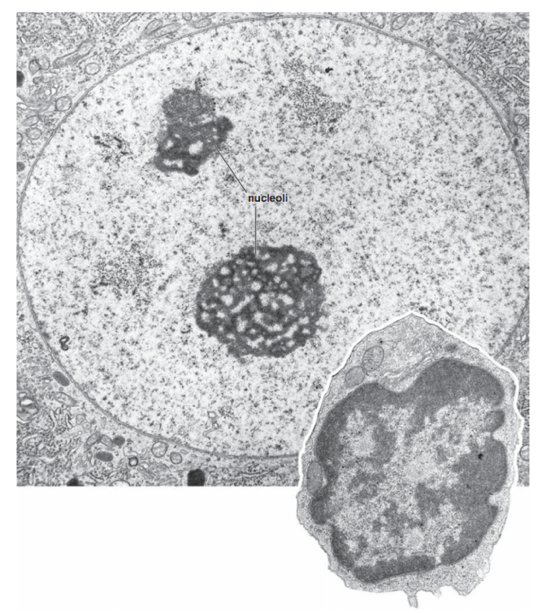
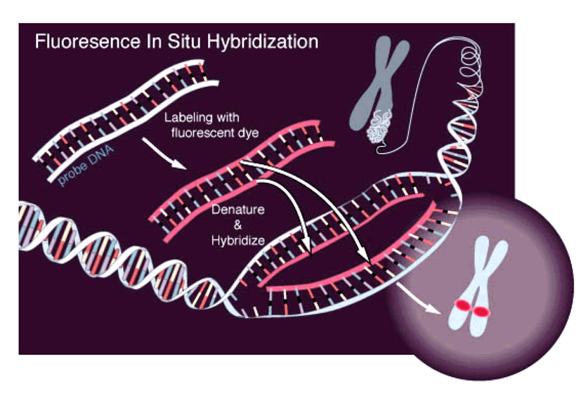
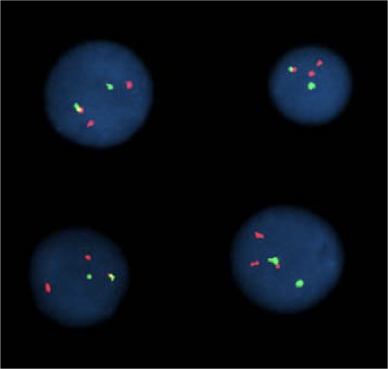
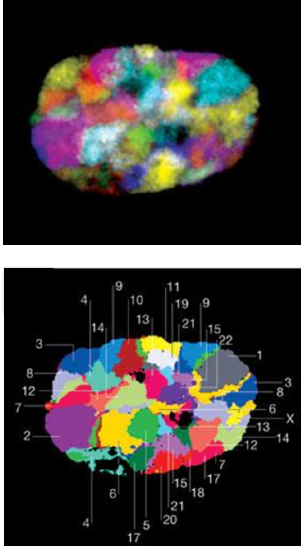
Microscopy:
Microscopy with fluorescing marks:
(FISH)
For 2 marks:
For multiple marks:
Chromatin conformation capture
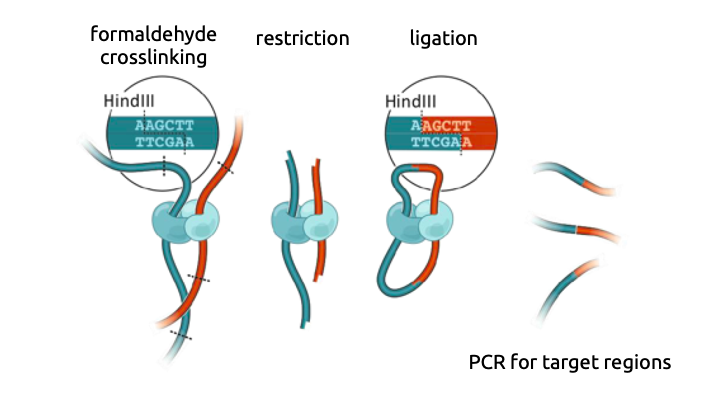
3C: Dekker et al. Science 2002
Hi-C
Viewpoint
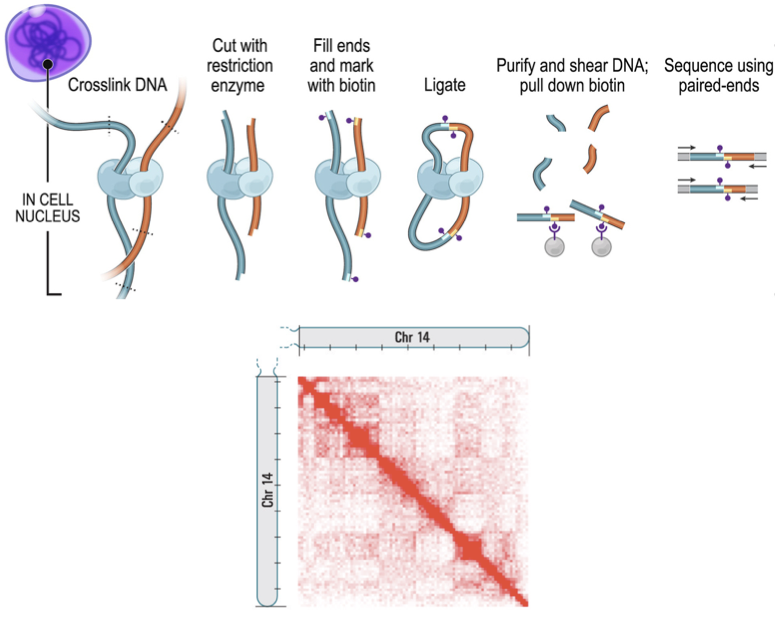
High-throughput chromosomes conformation capture:
DNA interactome map:
Lieberman-Aiden et al. Science 2009
Chromatin spatial structure
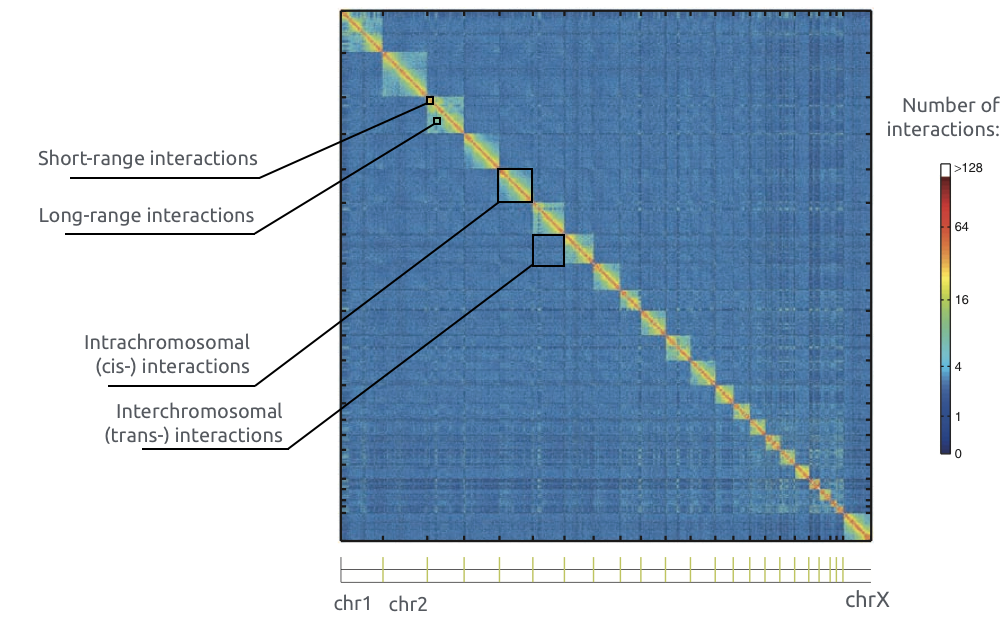
Adopted from Imakaev et al. Nature Methods 2012
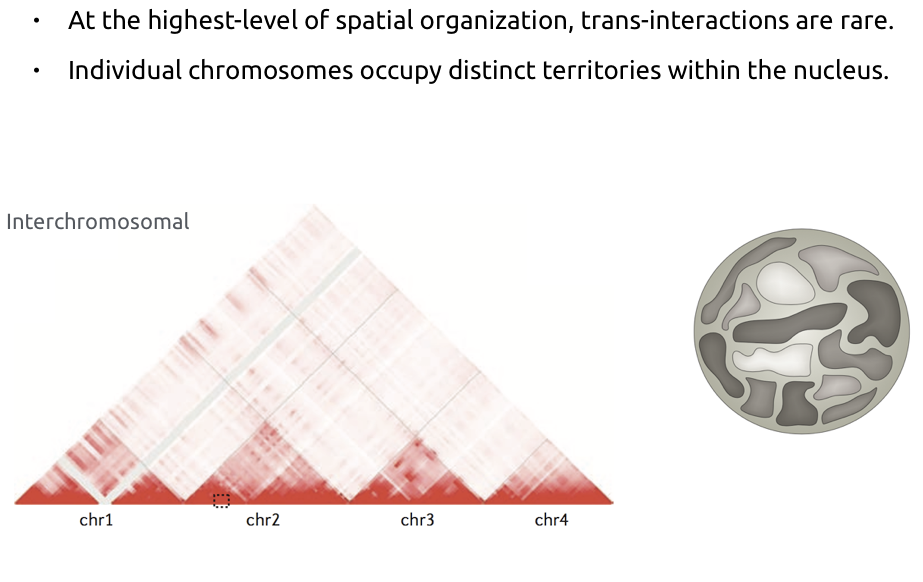
Chromosome territories
Bonev et al. Nature Reviews 2016
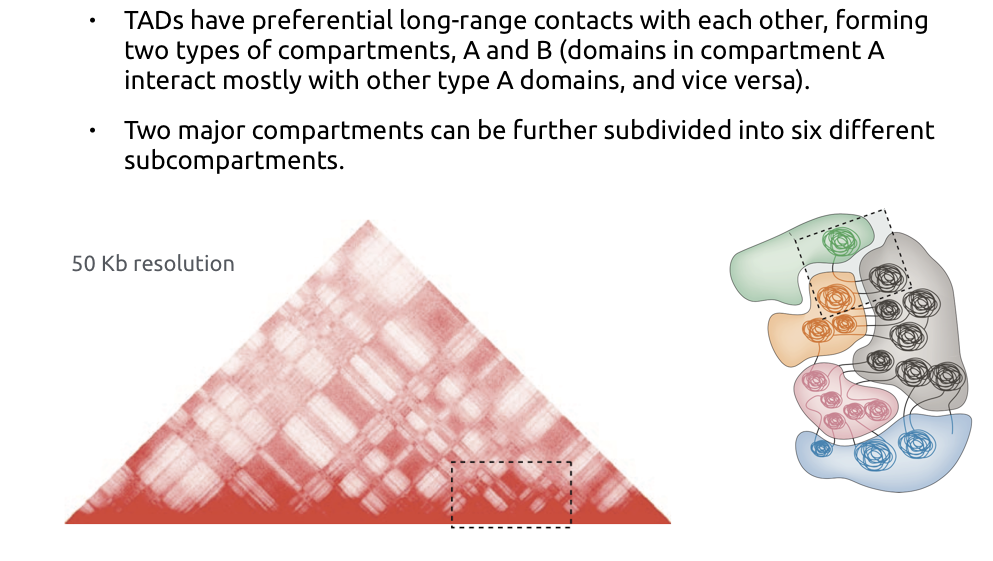
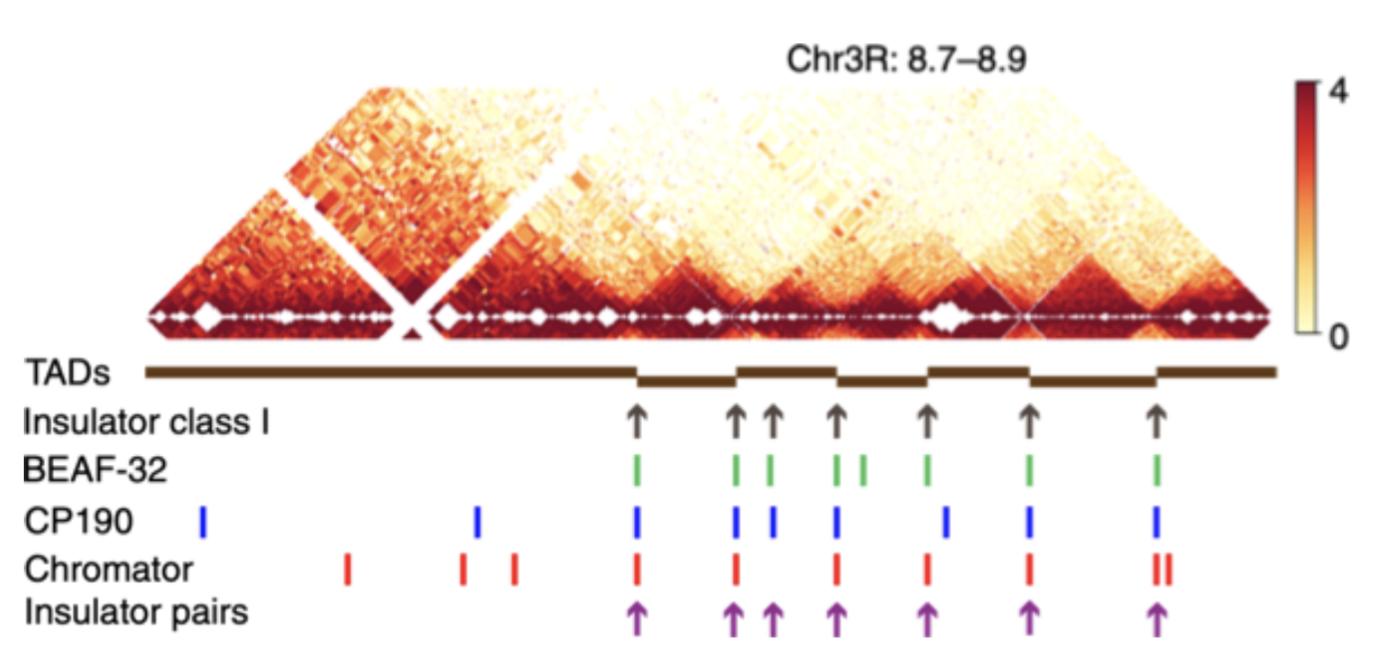
TADs
Bonev et al. Nature Reviews 2016

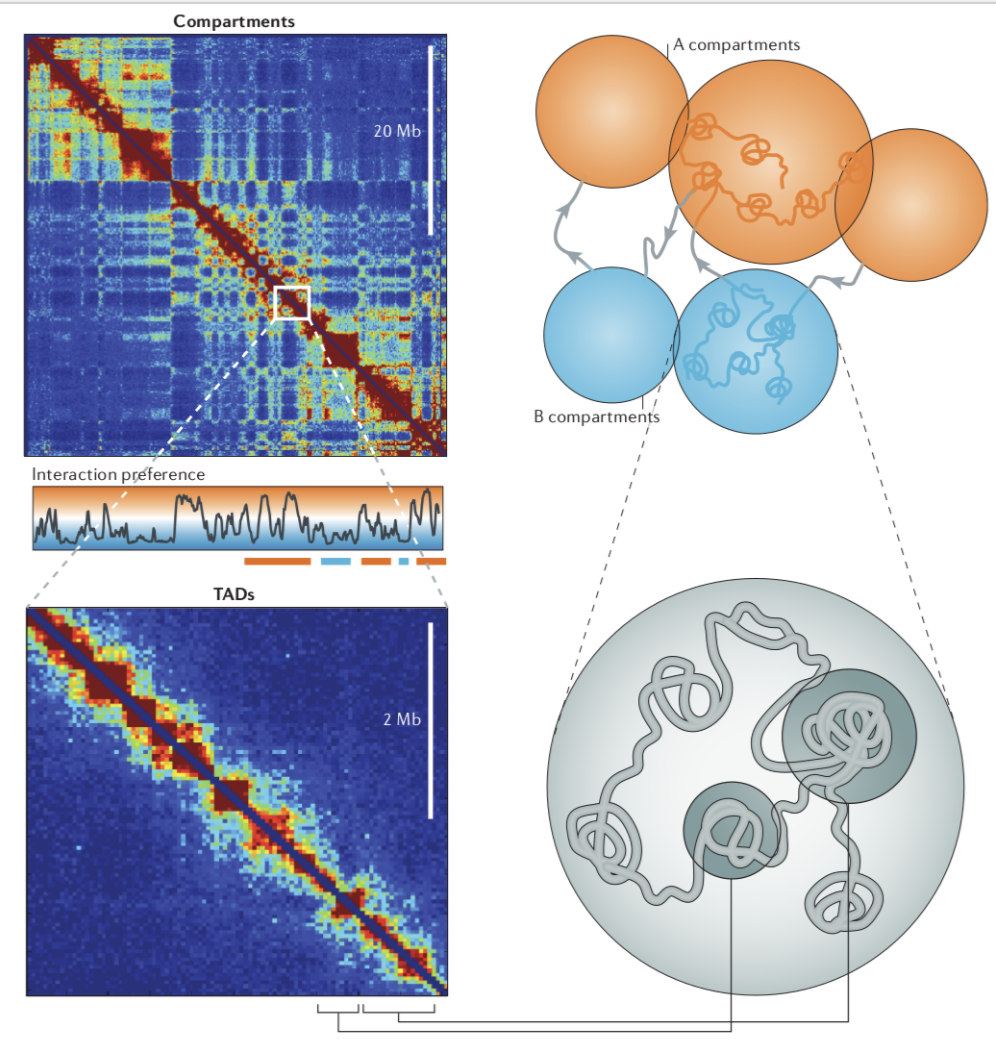
Compartments of chromatin
Bonev et al. Nature Reviews 2016
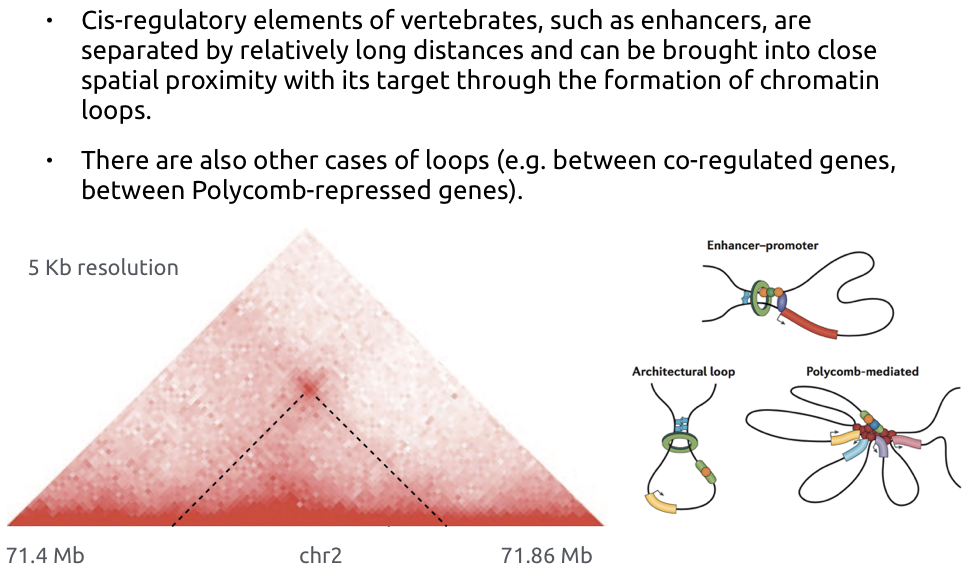
Loops of chromatin
Bonev et al. Nature Reviews 2016
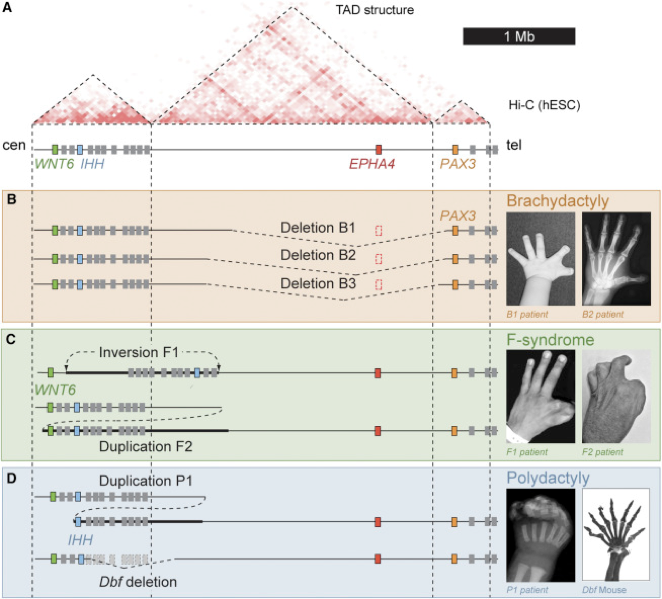
The importance of TADs and loops
Lupiáñez et al., Cell, 2015

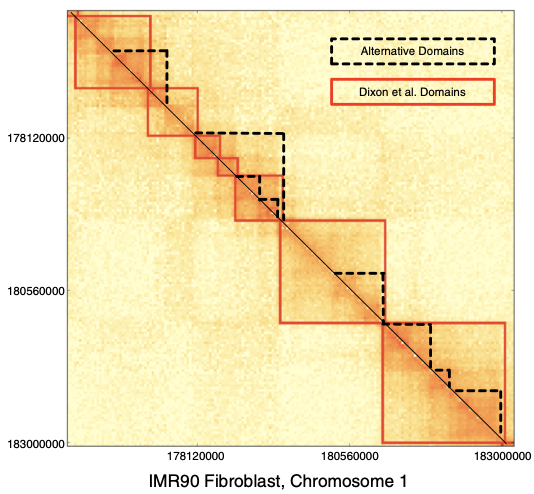
TADs
Filippova et al. Algorithms for Molecular Biology 2014
TADs are hierarchical, there is no single solution for the TAD calling problem:
- Armatus is one of the programs trying to solve this problem.
- Armatus is based on a dynamic programming algorithm that has an adjustable parameter.
TADs callers comparison
Forcato et al. Nature Methods 2017
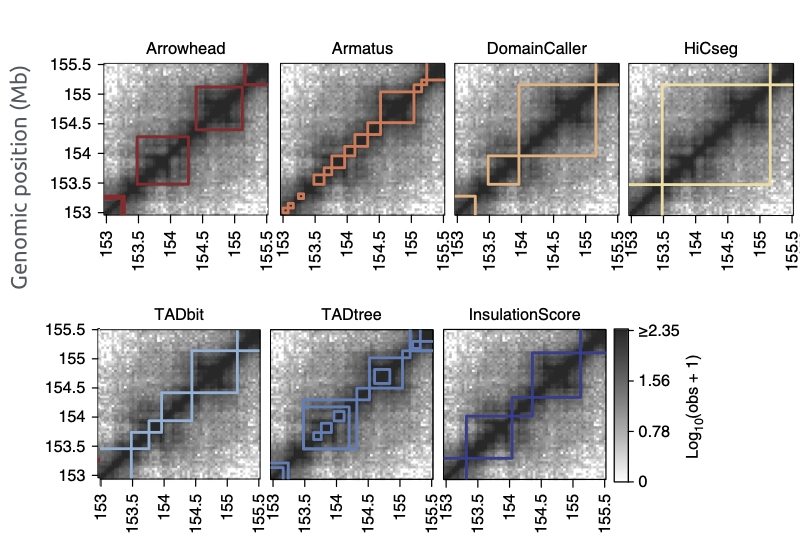
TADs callers comparison
based on Crane, 2015
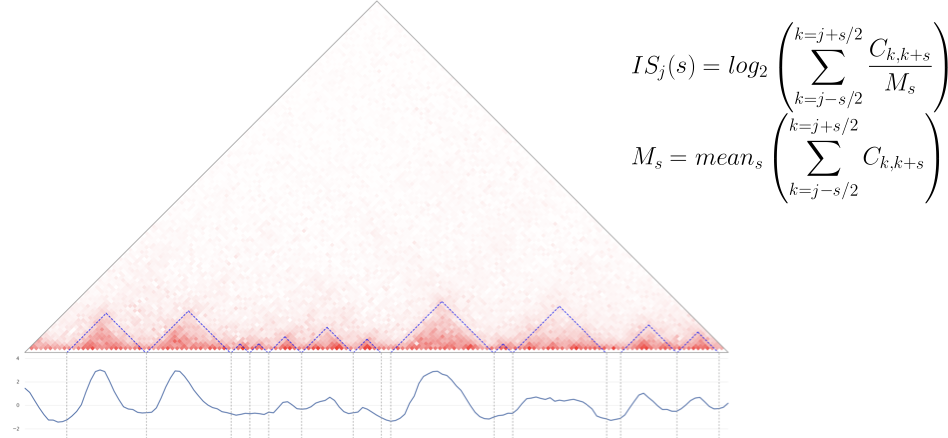
- Insulation score - is one of the simplest algorithms for TADs search:
- Calculate Insulation score for each genomic bin,
- Look for the local minima:
Functional annotation problem
Ulyanov, Khrameeva et al. Genome Research 2016
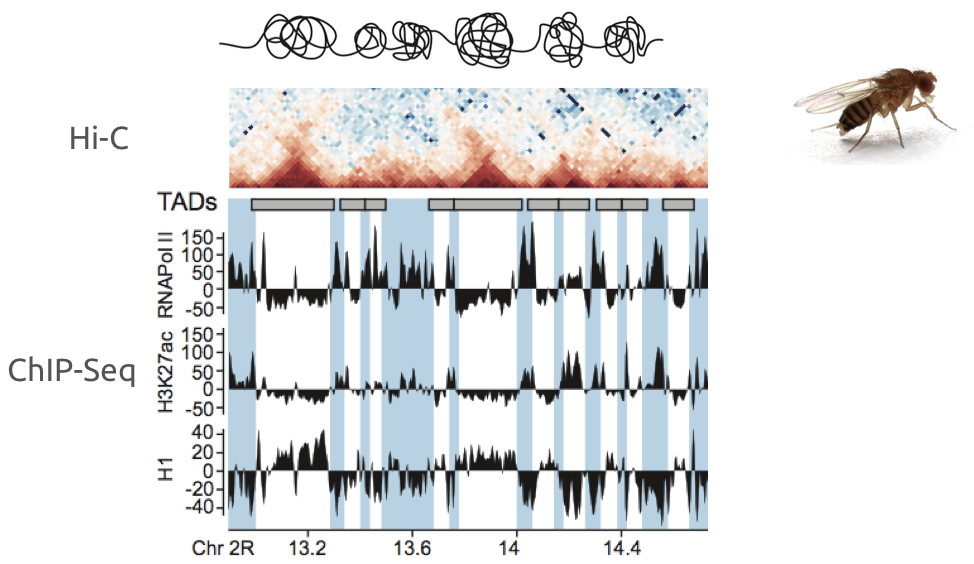
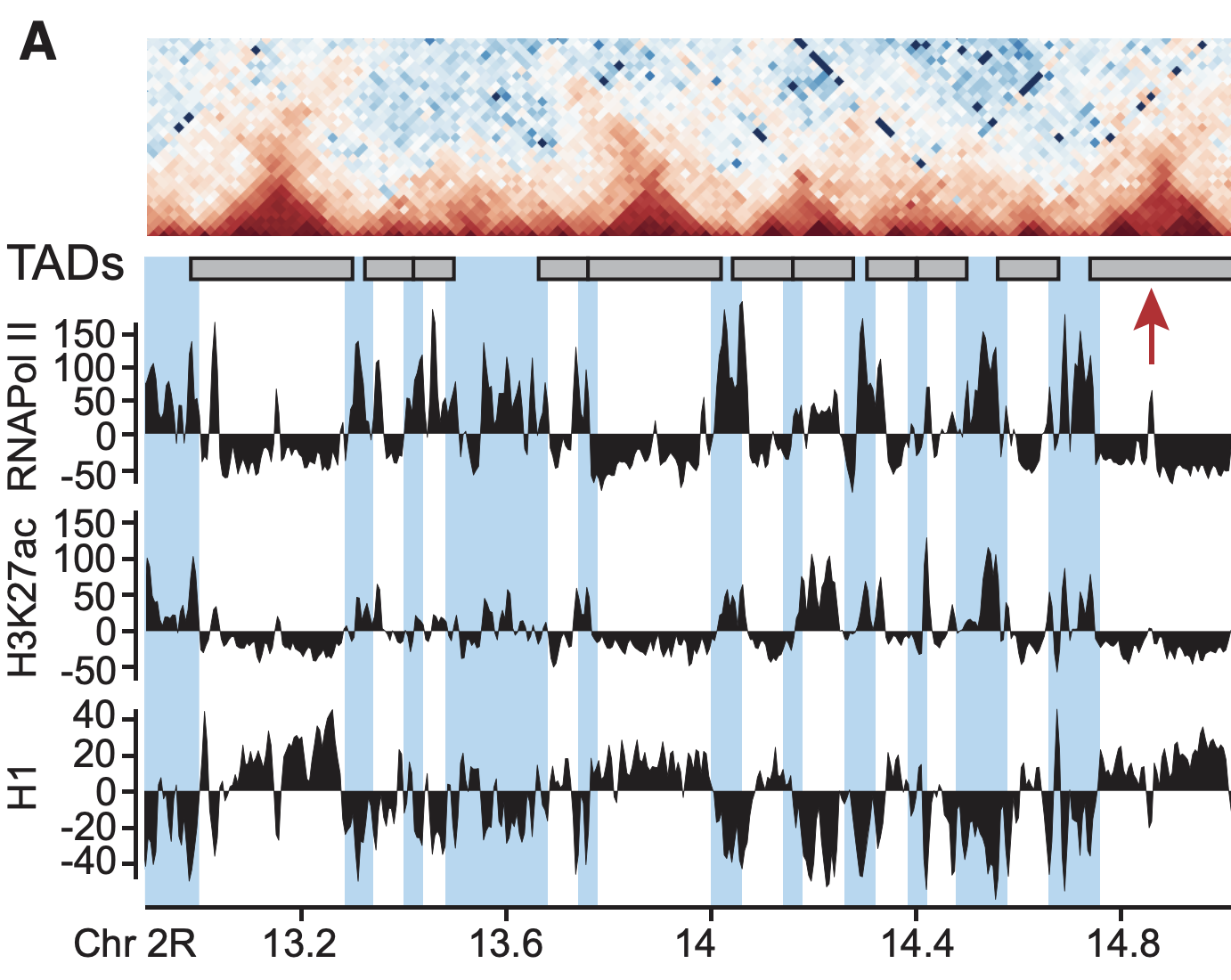
Example of association of complex datasets:
Stackup plot
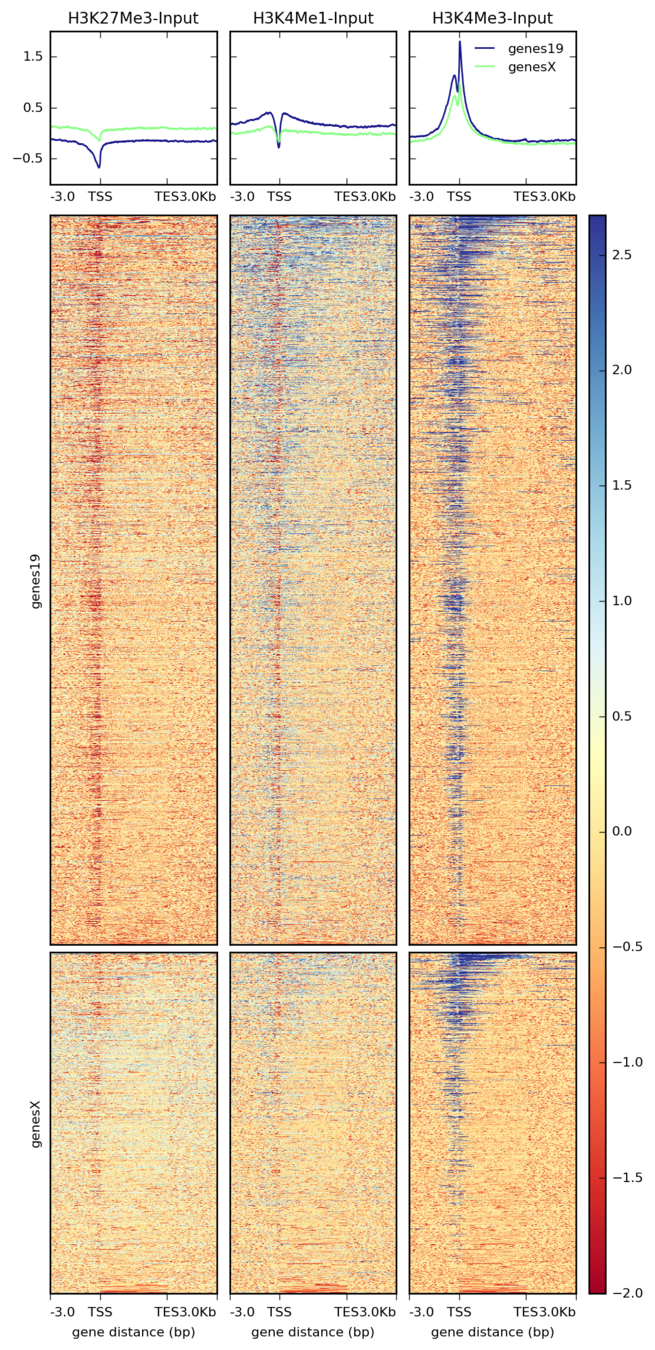
Stackup plot
Ulyanov, Khrameeva et al. Genome Research 2016
Plotting the profiles of enrichment around TAD boundaries (ChIP-Seq):
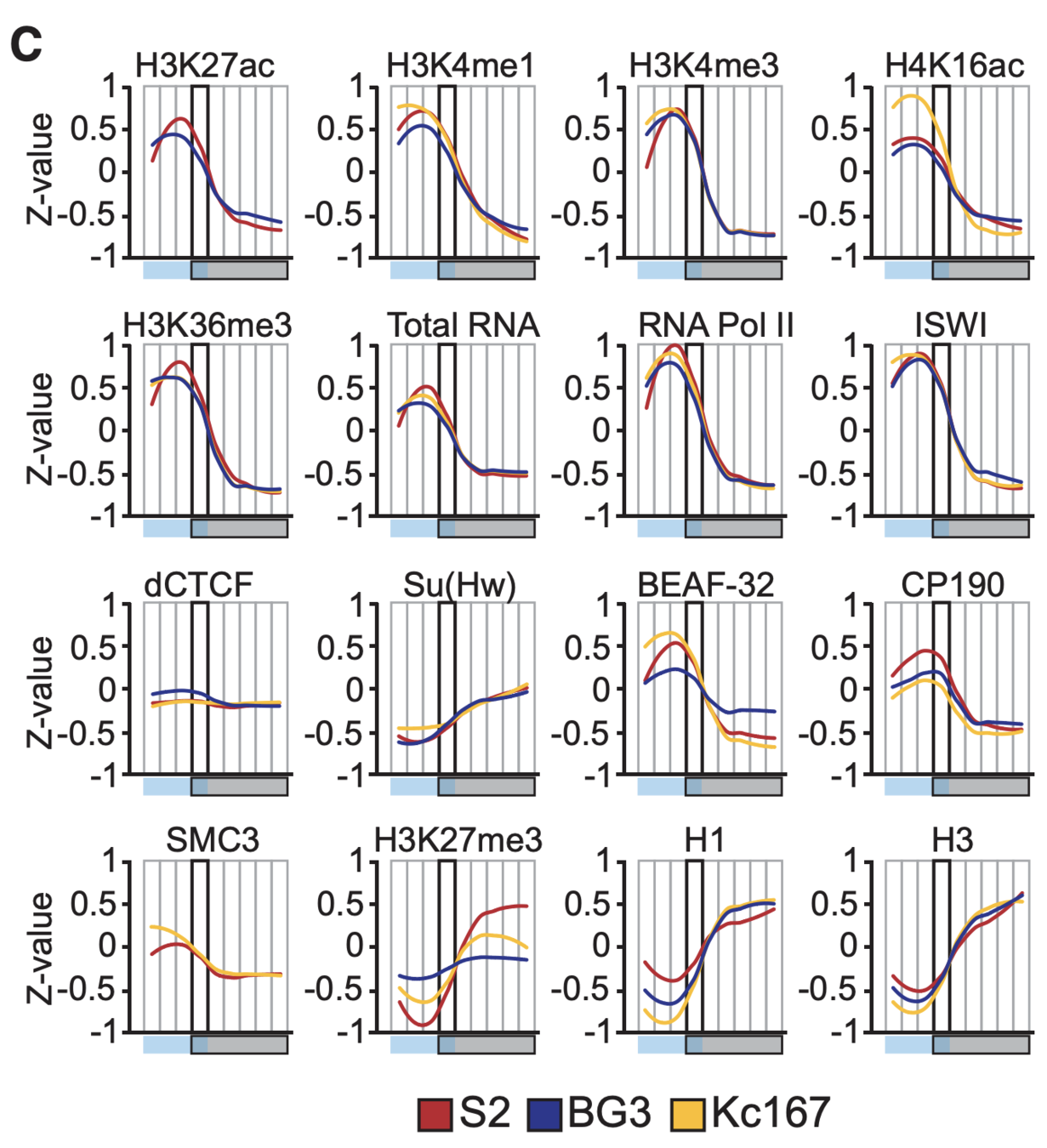
Functional annotation of the boundaries
Task:
- Download bigwig-файл for the factor you like from ENCODE. Good candidates are CTCF, Chriz, Su(Hw) and the one below.
Use the link for selection and download:
Get the resulting link:
https://www.encodeproject.org/files/ENCFF637XEP/@@download/ENCFF637XEP.bigWig
Download the annotation:
! wget https://www.encodeproject.org/files/ENCFF637XEP/@@download/ENCFF637XEP.bigWig
Download the package for work with annotations:
! pip install pybbi
Import the package:
import bbi
Create the stack:
stackup = bbi.stackup('ENCFF637XEP.bigWig',TADs_clustering.ch,TADs_clustering.bgn-20000,TADs_clustering.bgn+20000, bins=20)
Plot the stack:
import seaborn as sns sns.heatmap(stackup, cmap='RdBu_r')
and enrichment:
plt.plot(stackup.sum(axis=0))
Функциональная аннотация границ
Самостоятельные задачки:
- Скачать bigwig-файл для понравившегося фактора с ENCODE. Хорошие кандидаты: CTCF, Chriz, Su(Hw) и ниже.
Воспользуйтесь ссылкой для выбора и скачивания



Получите итоговую ссылку. Она должна иметь вид: https://www.encodeproject.org/files/ENCFF637XEP/@@download/ENCFF637XEP.bigWig
Скачаем аннотацию:
! wget https://www.encodeproject.org/files/ENCFF637XEP/@@download/ENCFF637XEP.bigWig
Установим пакет для работы с аннотациями:
! pip install pybbi
Импортируем библиотеку:
import bbi
Построим стек:
stackup = bbi.stackup('ENCFF637XEP.bigWig',TADs_clustering.ch,TADs_clustering.bgn-20000,TADs_clustering.bgn+20000, bins=20)
Нарисуем стек:
import seaborn as sns sns.heatmap(stackup, cmap='RdBu_r')
и обогащение:
plt.plot(stackup.sum(axis=0))
Theory of epigenetic coding
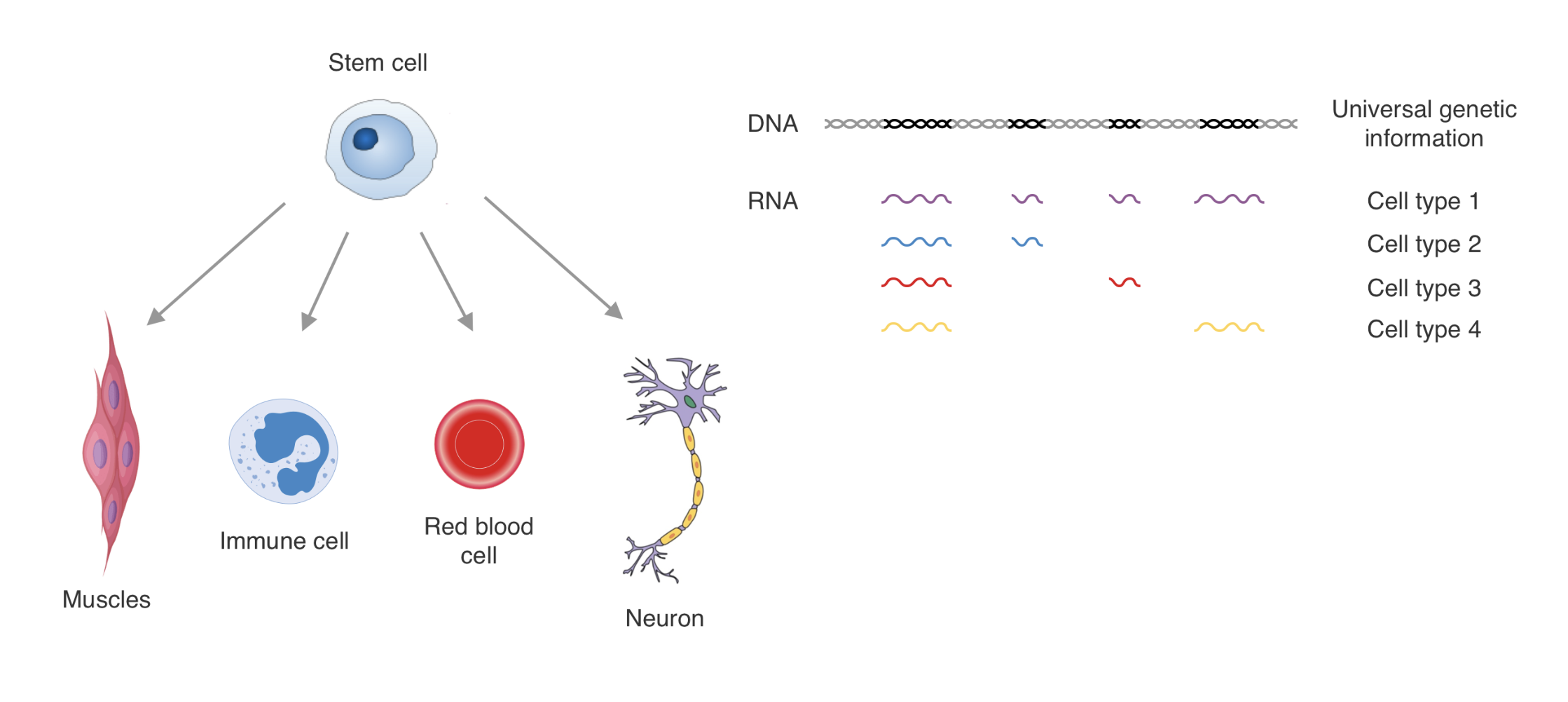
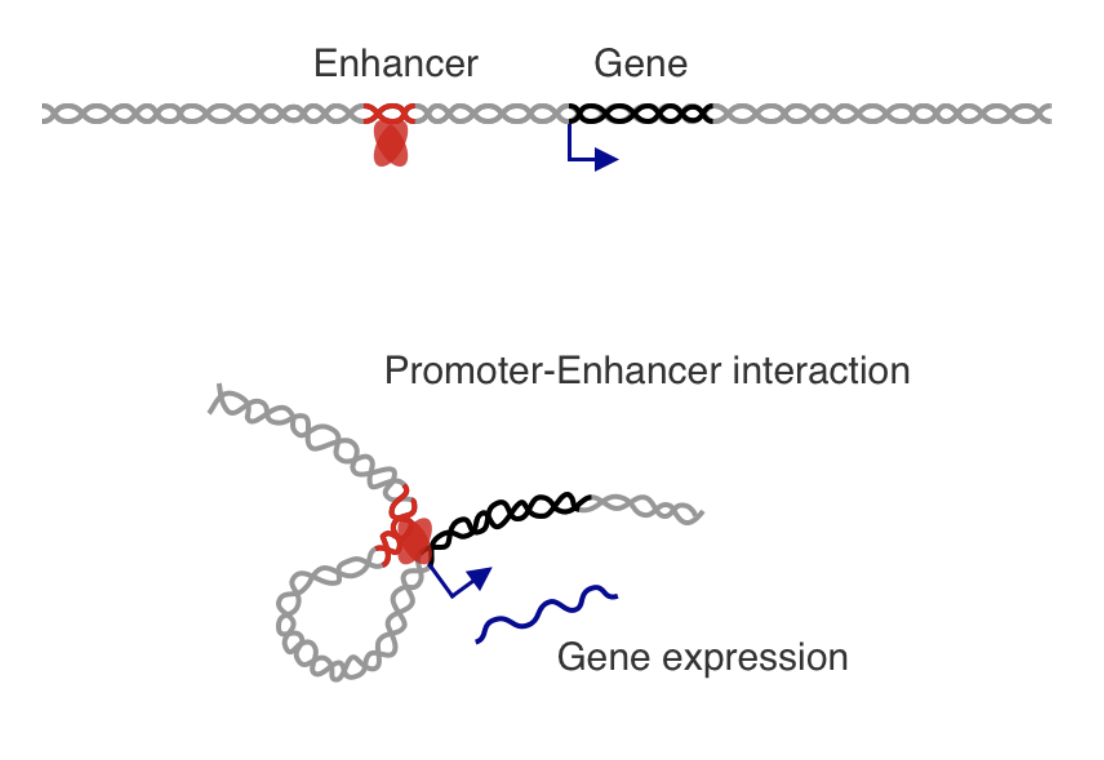
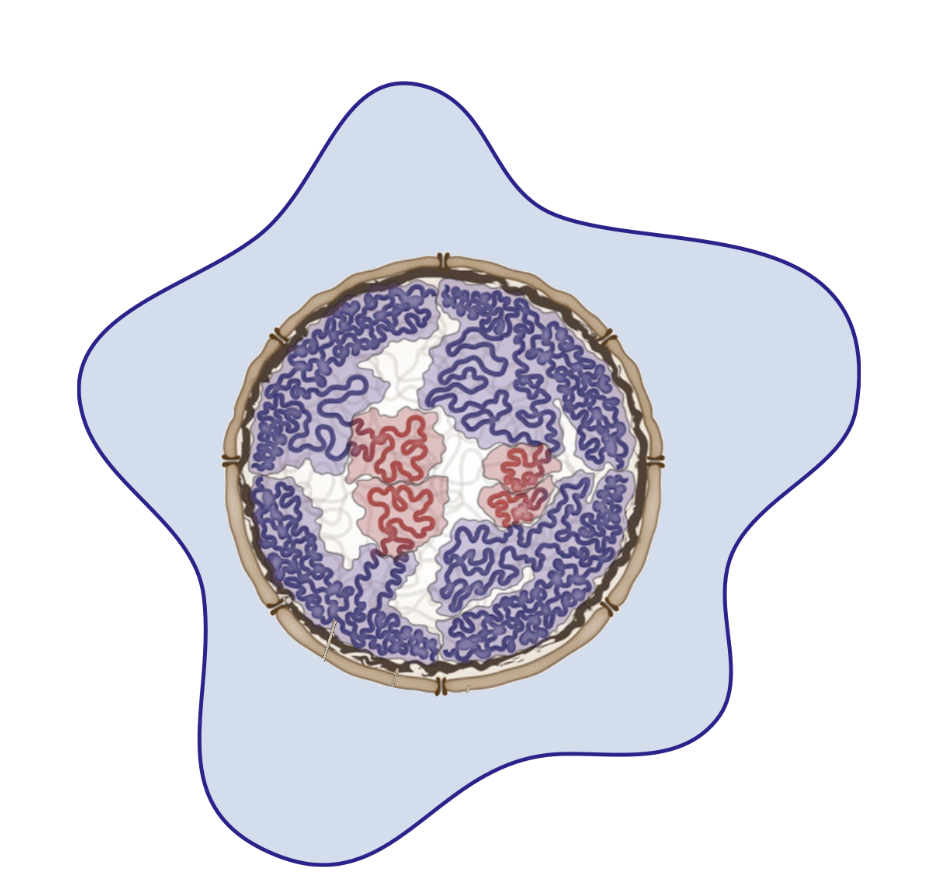
Chromatin in development
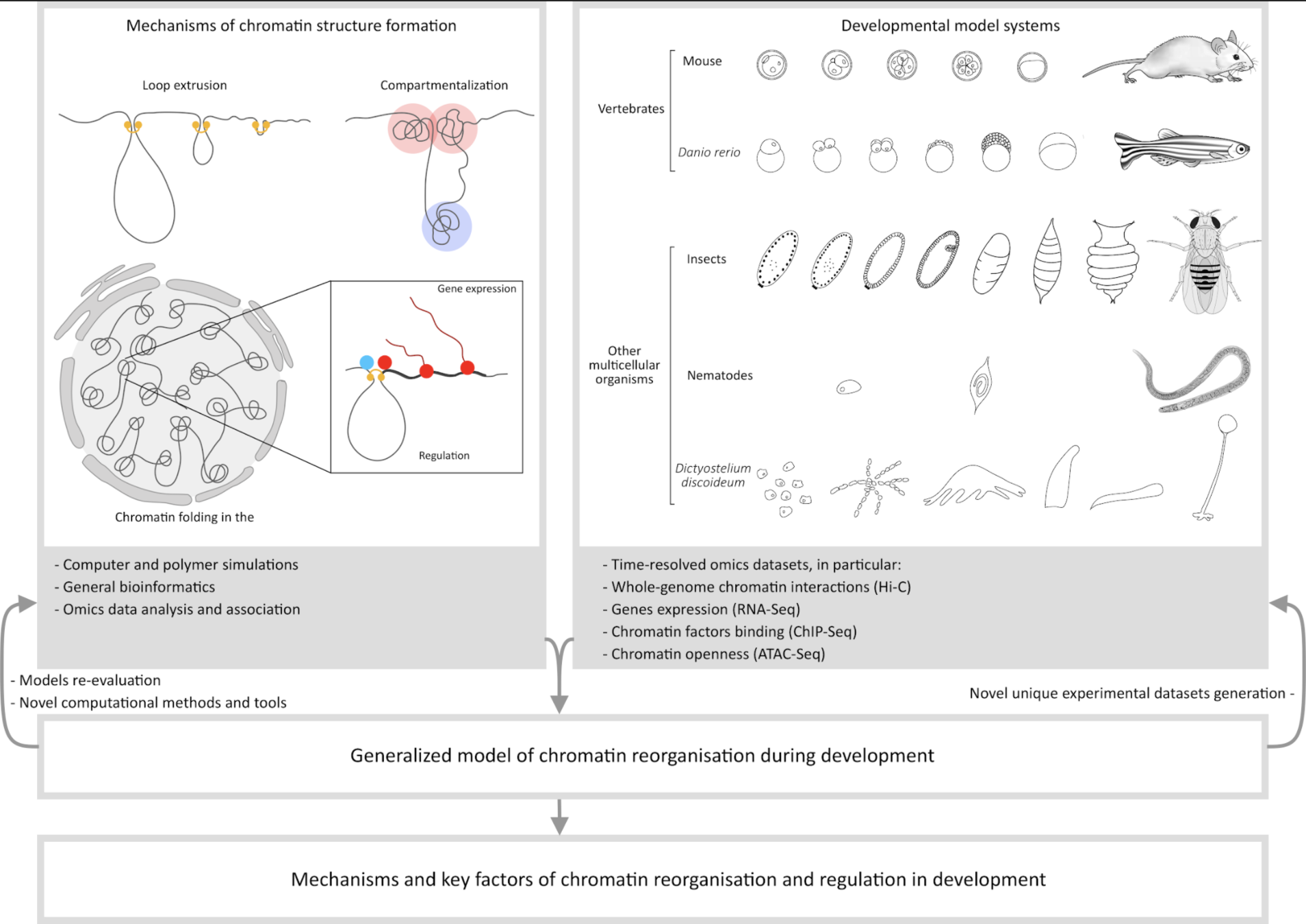
Chromatin as a graph
Graph in development

SMTB 2018
Bullet One
Bullet Two
Bullet Three
time-series-dna-graph
By agalicina
time-series-dna-graph
Welcome to time series lab at SMTB 2021
- 251



