Studying Paramecium's epigenetics with PacBio sequencing


Introduction
Transposable elements
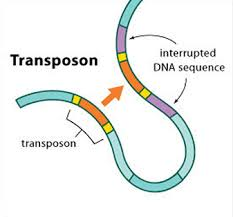






=
Present = Ruins that remained after the apocalypse(s)
Many strategies exist against TE
piRNA / siRNA
RNA guided Methylation of DNA
Histone modification
CRISPR / Restriction enzymes
Hypermutation (neurospora crassa)
Random excision
RNA decays against non-sens ORFs
...
The ciliates: a specific case
P. tetraurelia: Genomic architecture

- Eucaryote, Ciliate
- 3 nuclei:
- 2xMIC nuclei (2n)
- Reproduction
- Contains:
- TE & IES
- 1xMAC nucleus (up to 800n)
- Transcription
- Partial MIC
- 2xMIC nuclei (2n)
Sexual processes
2 sexual processes:
A) Autogamy
B) Conjugation

Everytime : Karyogamy of 2 haploid MIC
A new MAC is formed according to the new MIC, with the proteins and the leading of the old MAC
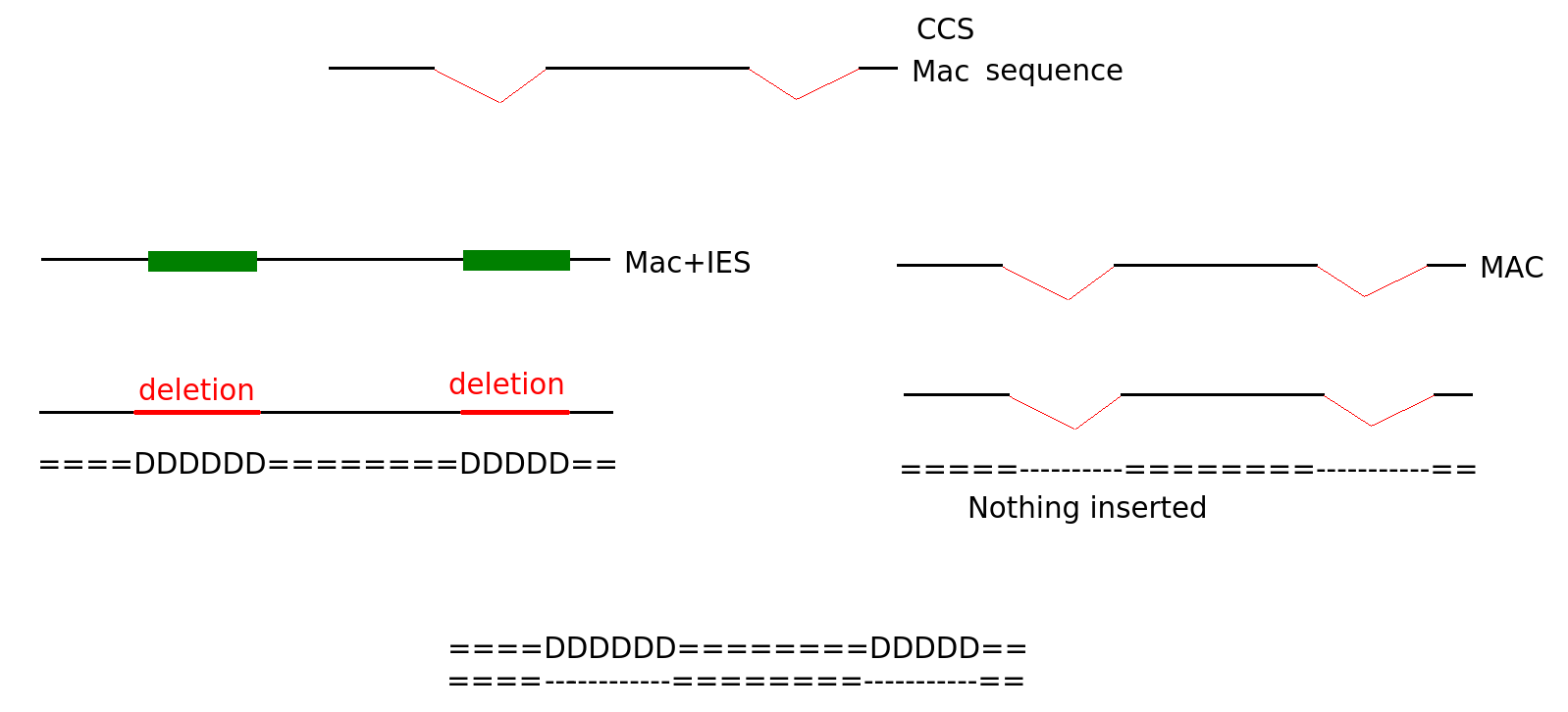
Transposable elements and IES are removed during the process
IES excision in the new MAC
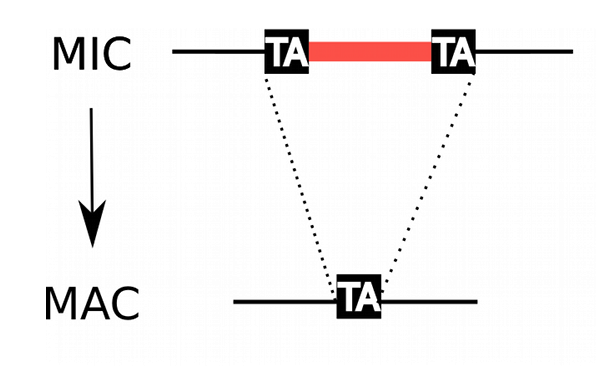
TE/IES: suppressed in the MAC (transcription OK)
--> Avoids the negative effect of TE and IES
~99% IES have TA-boundary
No other consensus sequence, unlike other ciliates
Lots of IES inside coding sequences
~ 100% PGM-dependant excision


PiggyMac
IES
Excision of IES: ScanRNA pathway
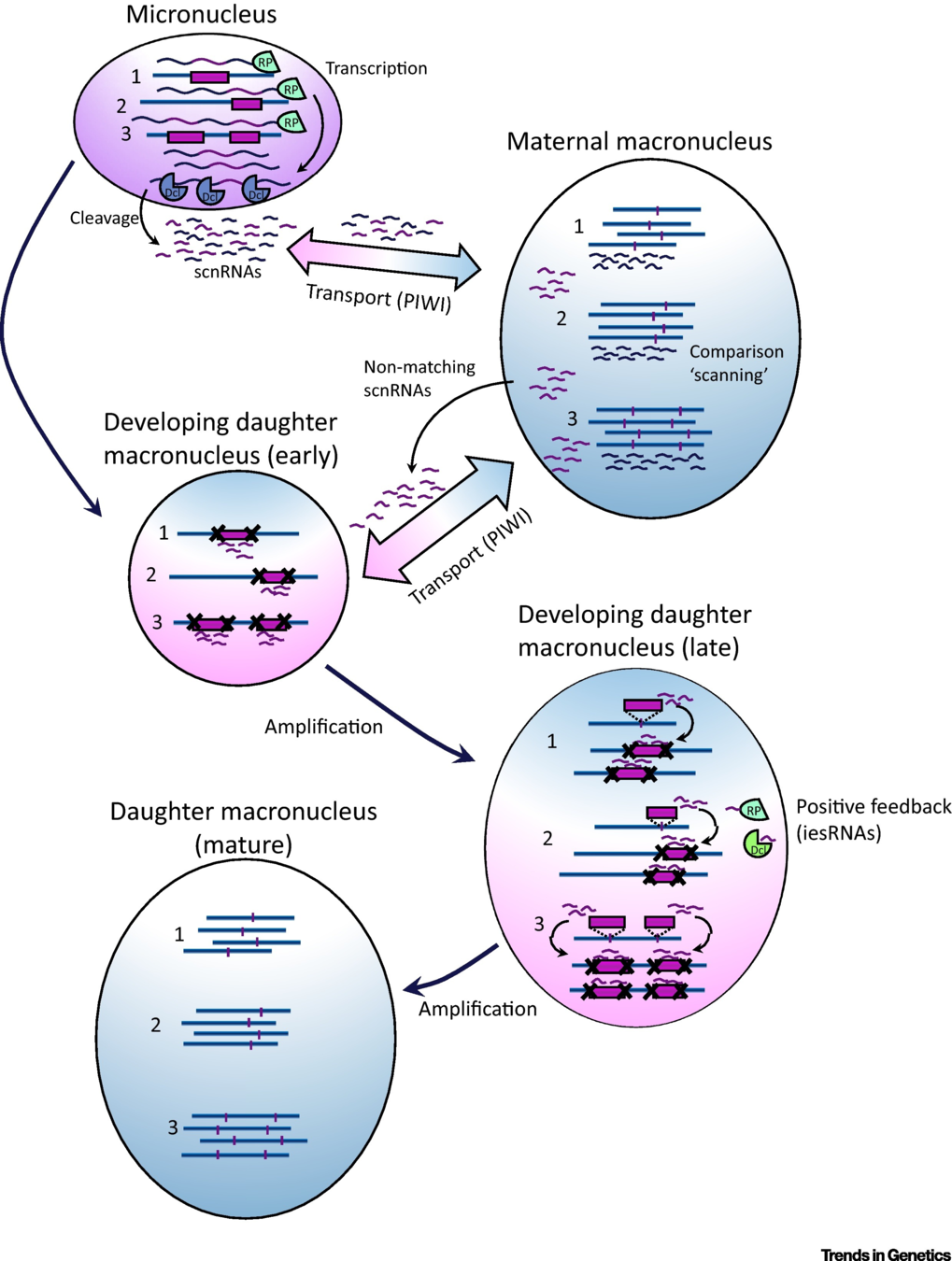
- SCAN-RNA --> Excision of 60% of IES max (shown by DICER-like silencing)
- Piwi shuttle
- What about the 40% remaining ?
DNA Methylation ?
So, recently in Eric's lab...
Modified bases play an important role in:
- Procaryote's DNA/RNA
- Eucaryote's RNA/DNA
Lots of "orphan-MTases"
Remains highly misunderstood
Seeking 6-mA recognition domain
Protein identification
RNA silencing

Death of
progeny after autogamy/conjugation
--> Role in IES excision ?
Currently being studied
Hypothesis
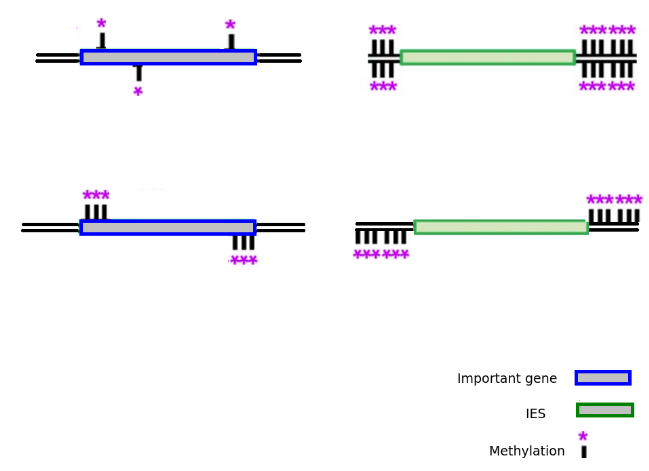
Goals
Sequence the methylation
Sort MIC and MAC DNA data
Compare, especially on the IES boundaries
Materials & Methods
PacBio sequencing meets all expectations
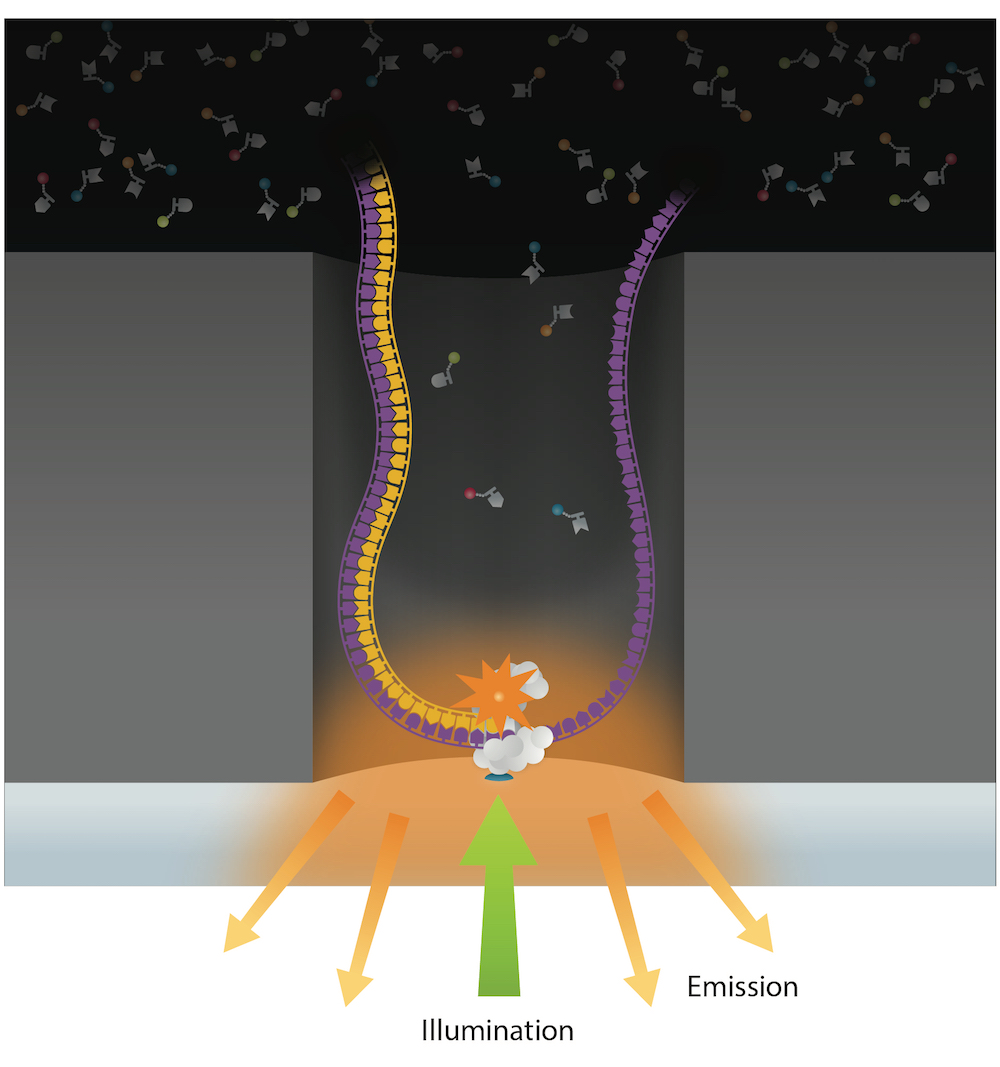
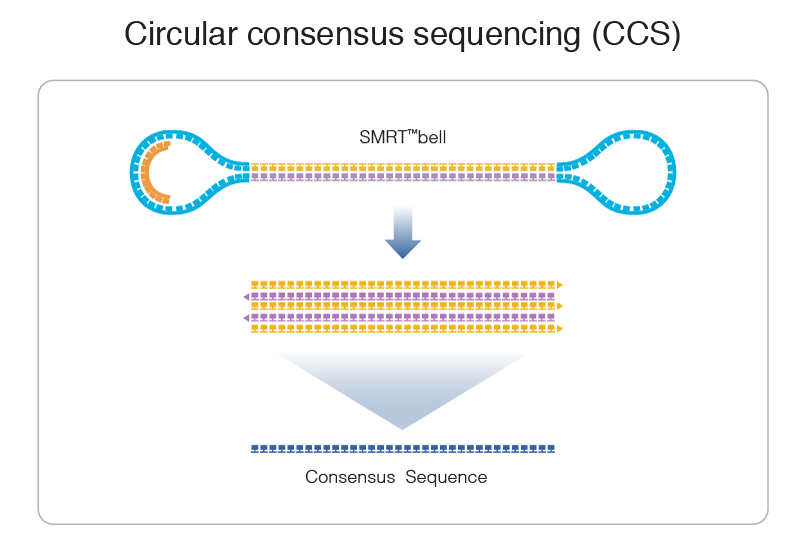
99% accuracy
Max
75% accuracy
Methylation analysis needs local > 25X
Trained model (ML) allows detection of suspect downturns of polymerase (function of the -3/+8 nt context) --> IPD are captured
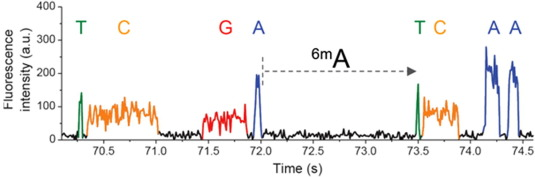
SMRT tracks 3 DNA methylation
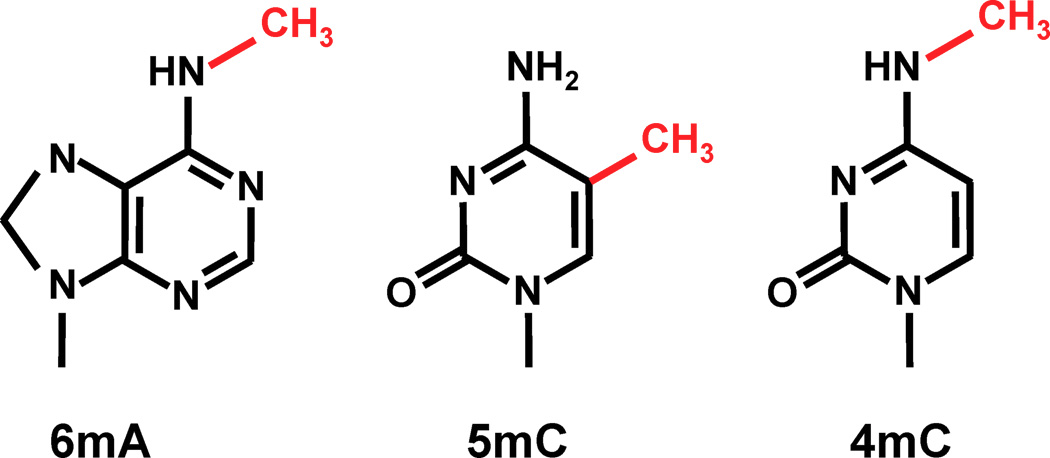
<-- 3 known most frequent
methylations in DNA
6mA is the most suspected one in Paramecium
Sorting the sequences
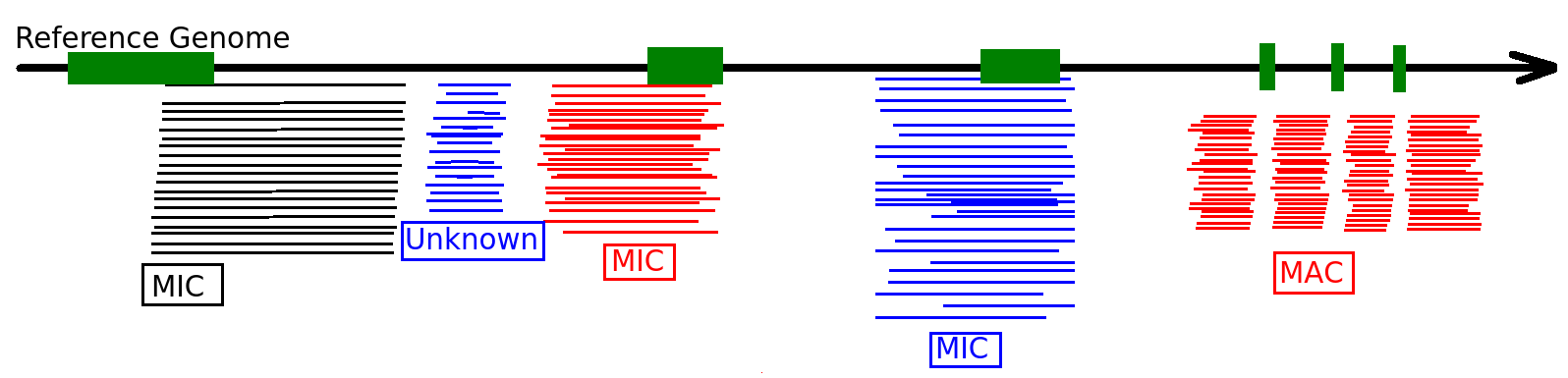
Unknown = Mac Destinated Sequence (MDS)
Available data
PacBio sequencing
Wild type:
- Vegetative Cell (HTVEG)
- Post autogamic cells 2h (HT2)
- Post autogamic cells 6h (HT6)
Silencing of methylase candidates:
- Si/MAB
- Si/MT2
- Si/MT1A-1B
- Si/MT1A-1B-2
- Si/NM4-9-10
- Si/NM9-10
- Si/NM4
Other:
SiPGM
Previously ...
Sorting DONE
Hacking for Single-molecule (IPD, capping) DONE
Methylation analysis (production of outputs) DONE
Work on cutoffs, scores, GMM... DONE
AT and TA, various score stats DONE
Motif analysis DONE
MDS To-do
Transcription start site To-do
IES scRNA VS undepending To-do
A few flashbacks on the results
Diffential mapping
Sequences that come from MAC
Sorting stats
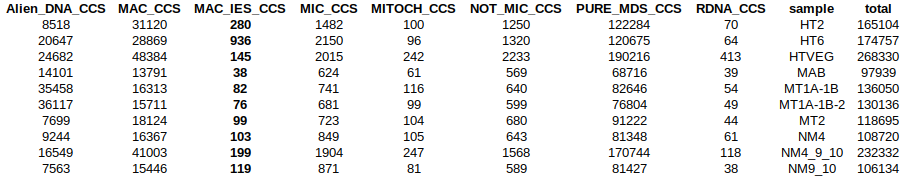
rdna ??
mito ??
Score distributions: The case of the adenines
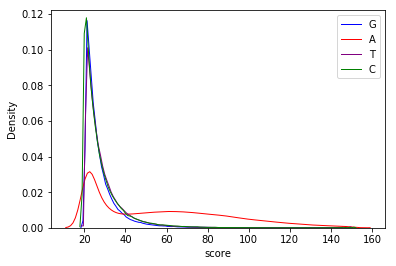
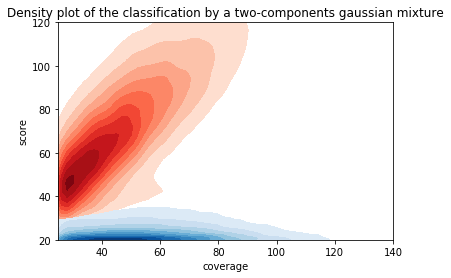
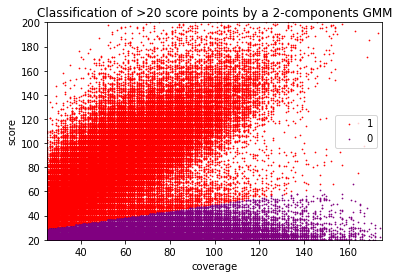
Modifications in AT/TA
- 95% AT
- Same MIC/MAC in AT
- No difference between experiments
- ~75% methylated symetrically
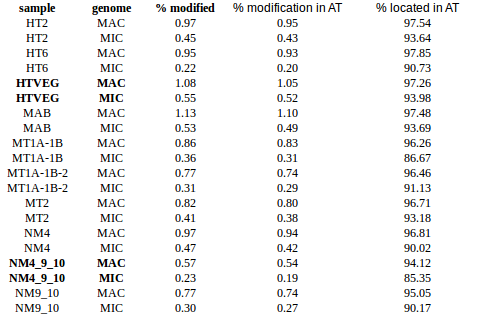
Modest variations in the silencing
Modifications out AT
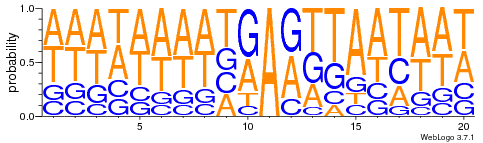
What interests us most
We didn't find no difference between experiments
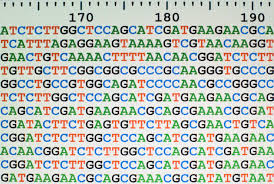
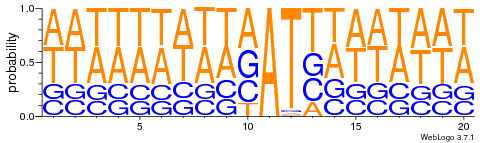
FIndings in cytosines

Logo from HTVEG MAC
Identical everywhere
I fixed some problems inbetween
Sorting was wrong --> Only MAC data can be trusted FIXED
RDNA and mito are back <3
Missing RDNA and Mito --> Pb for southwesternblot FIXED
Mis-excised IES in the MAC --> False IESs FIXED
Mis-estimation of % because of missing MDS --> FIXED
Scan-RNA handling --> SOON
GMM --> Unsatisfactory ONGOING
Slow computation ??
Dataset size ??
All stats and motifs must be done again now that the sorting is fixed
Reassuring findings
- Consistency with Gif's Lab on our results
- Consistency with new litterature on batched-SMRT
- Consistency with new evidences on ocytrichia and tetrahymena
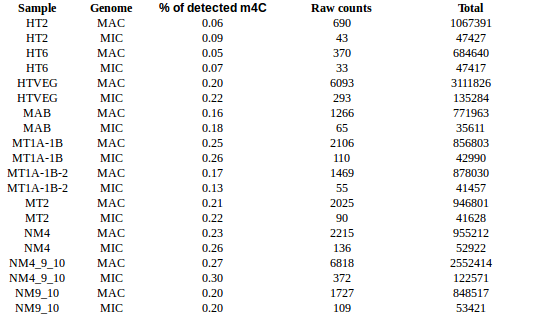
- We found 6mA in the MIC and we have exclusivity for that
- No one has our resolution for the moment
- Homologs of our silencing were found to be relevant in other ciliates this year
- Experience acquired from errors this year...
- m4C in MIC (and MAC !) absent at T2 and T6 would be a real scoop for eucaryotes
Tasks for the near future
percentages, motifs analysis, subpercentages, multiple score thresholds testing... Redo what I've done before

And to conclude...

SMRT - Sept 2019
By biocompibens
SMRT - Sept 2019
Lab meeting - 19/06/18
- 97



