Self, Non-self
ID-spoofing
Guillaume DELEVOYE
Lab Meeting 09/04/19

Everything is unique

Everyone is unique





What makes you unique ?
Blood type
Genetic variants
DNA
Measurable attributes
Personal history
Personnality
Tastes
Health status
Name
Date of birth
Parents
Combination makes you unique
Blood type
Genetic variants
DNA
Measurable attributes
Personal history
Personnality
Tastes
Health status
Name
Date of birth
Parents
A x B x C x D x E x F ...
Spoofing 101


Security checks are efficient, quick, and therefore rely on checking proxies to your identity
ID Card
Whitelist
Appearance
Password
What specification for a security system ?
- Adaptation to fraud
- Recognition of danger VS recognition of safety
- Contextualisation
- Tolerance
So how would the nature do ?


Comment fait la nature pour:
1 - Faire la même chose que nous ?
2 - Le faire aussi bien mieux que nous ?
Elle fait peut-être exactement comme à peine différemment de nous

In practice
How would the nature do ? (Immune System)
- ID Card
- Whitelist
- Blacklist
- Adaptation to fraud
- Adaptation to the new
- Threat-depending analysis
- Recognition of danger
- Recognition of safety
- Contextualisation
- Tolerance
- Memory
- Multiple checkings
- Tattooing
- Communication between agents
Natural killer, missing self immunuity
HLA system
PRR
Th1 cells
Antibody selection
Cell proliferation
PAMP
CMH/HLA-DR
Inflammation
Dendritic Cells, Anergic T cells &
Isotypical conversion, memory B Cell
T Cells interactions
Opsonisation
Hormonal stress
Self, non-self, ID-spoofing
How is it related to my work
Pharm D Thesis:
- Donor-recipient compatibility in graft: Hacking the stem cell's ID-card
PhD Thesis:
- How the genetical non-self is recognized in Paramecium

How the genetical non-self is recognized in Paramecium






=
Present = Ruins that remained after the apocalypse(s)
Apocalypse now

Transposable elements
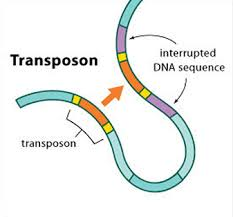
Horizontal transfer of TE
- TE are considered as "sharing an ancestor with viruses"
- They could transfer horizontally by their own or through pathogens, pollinators, symbiosis, plasmids...

... And many others
Autonomous copy/paste + horizontal transfer = Global Invasion
TE = Chaotic invasion ? Yes !
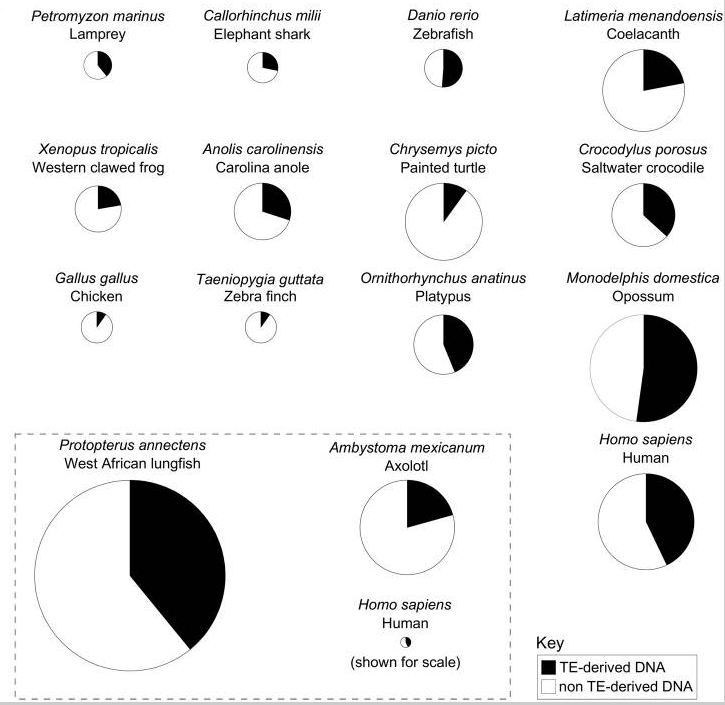
Genome Biol Evol. 2017
"Evolution and Diversity of Transposable Elements in Vertebrate Genomes" (Cibele G.)

Extreme case 1)
Maize

Up to 86%
He dead ?
Extreme case 2)
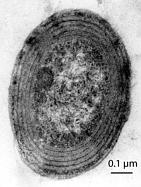
- Prokaryote
- One of the smallest genome ever sequenced
- No TE, no transposase
- Only known exception to day

Hypothesis (none proved):
1 - Very high multiplication rate + Very high vulerability to TE
2 - Defense mechanism so great it's not affected
Prochlorococcus marinus SS120
The selfish gene - 1976


- Finalist/Anthropomorphist analogy: "The DNA is selfish and only 'wants' to reproduce itself" --> Parasitic DNA
- With limited resources, the best DNA replicators "win"
- The evolution and natural selection could be better described by "DNA replicators" rather than species






=
If it survived
It should have been adapted to TE
Many strategies exist against TE
piRNA, siRNA
RNA Guided Methylation of DNA
Histone modification
CNVs
Hypermutation (neurospora crassa)
Random excision
RNA decays against non-sens ORFs
...
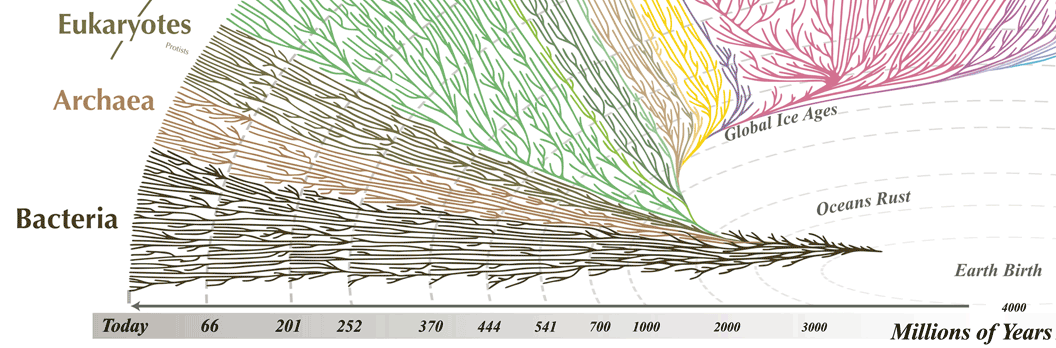
The global story remains a mystery
- A wide range of strategies exist
- Eucaryotes, procaryotes and archea have very different mechanisms
- This suggest a progressive, separate, even constant adaptation
The ciliates: a specific case
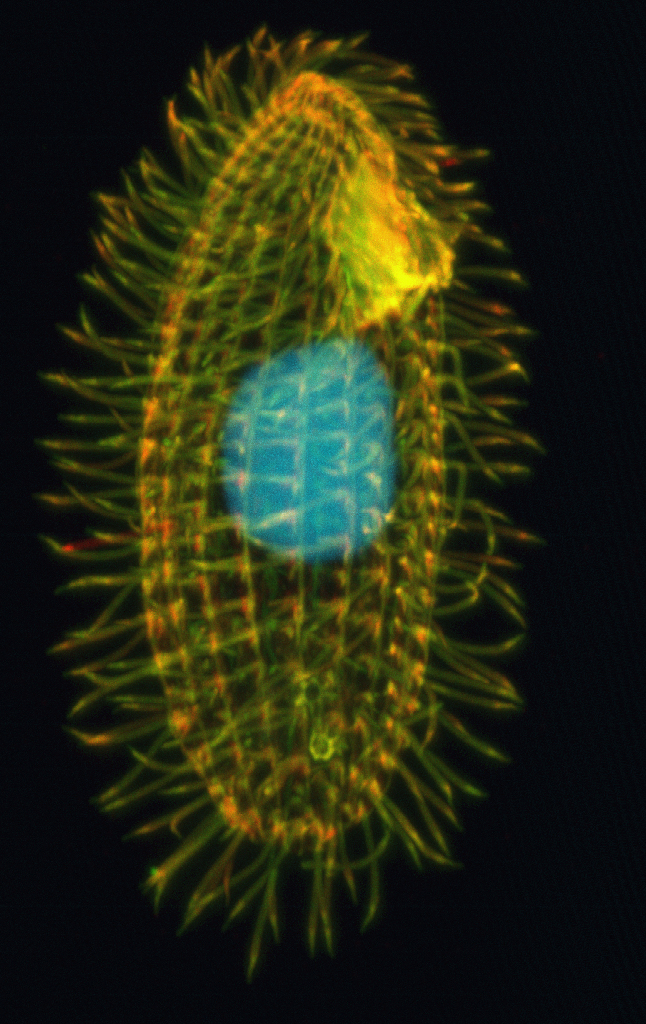
Paramecium tetraurelia

- Eucaryote, Ciliate
- 3 nuclei:
- 2xMIC nuclei (2n)
- Reproduction
- 1xMAC nucleus (up to 800n)
- Transcription
- Partial MIC
- 2xMIC nuclei (2n)
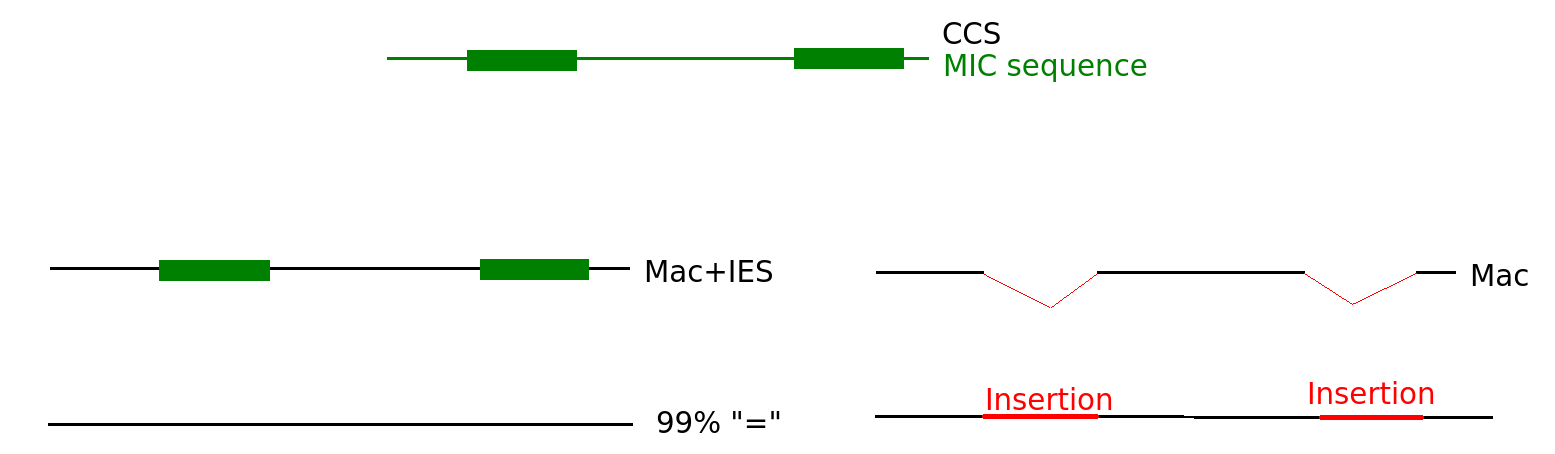
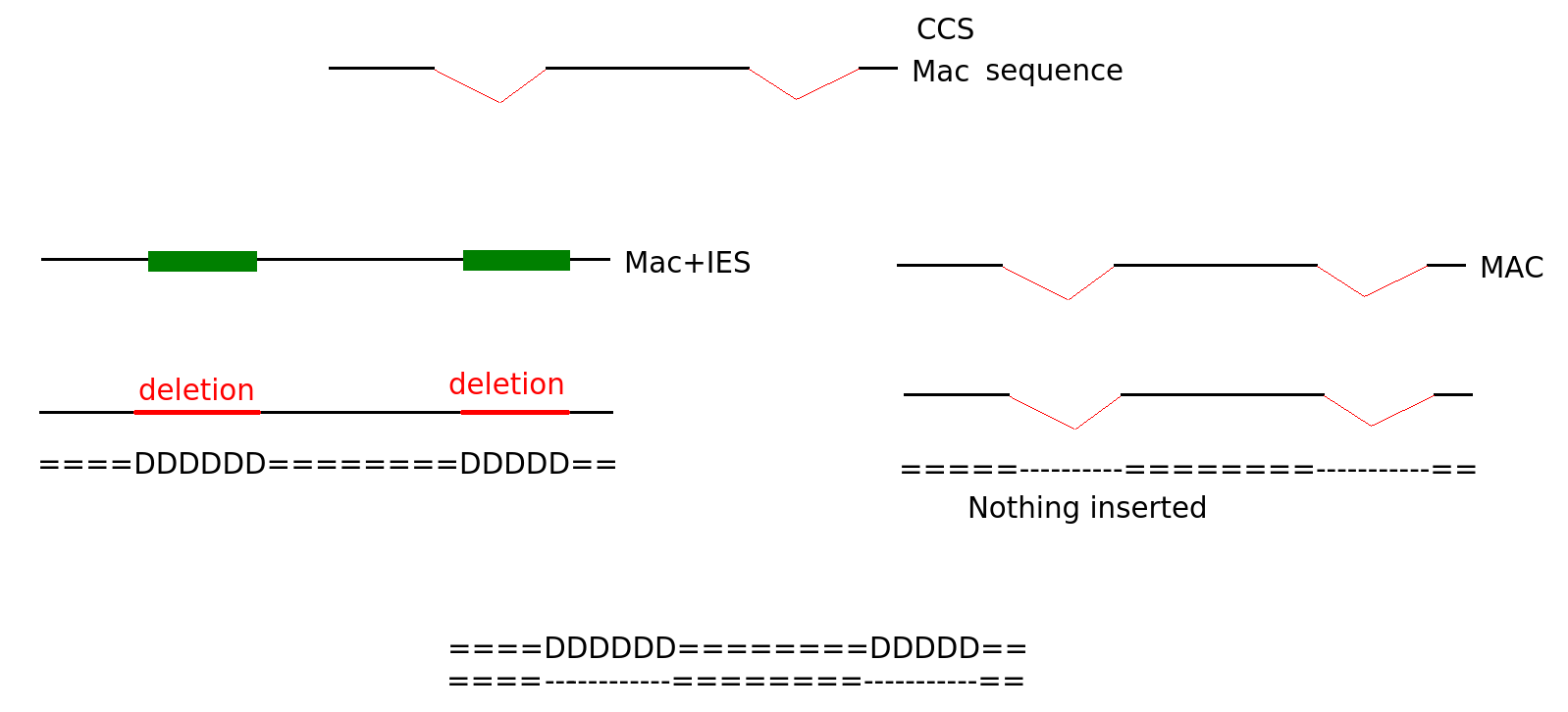
MIC/MAC Differenciation
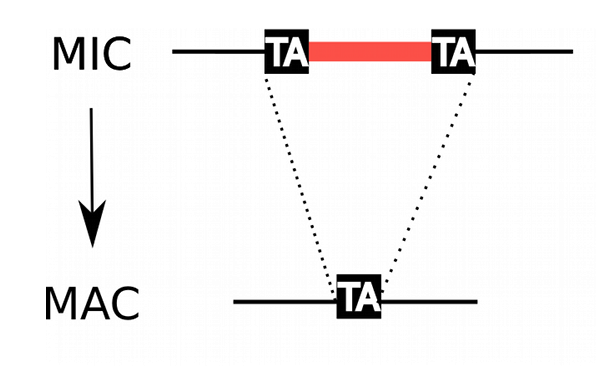
Transposable elements are suppressed in the MAC, where the transcription occurs
Avoids the negative effect of TE


PiggyMac
IES
In 2018, question is...
...How does the cell know where to cut ?

And even more than that
-
WGD potentially implies a lot of ohnologous TE
-
How would the cell still recognize them over the time ?
Genetic drift...
Answer:
It's not fully understood
Hypothesis
Please cut me !
Don't cut me !
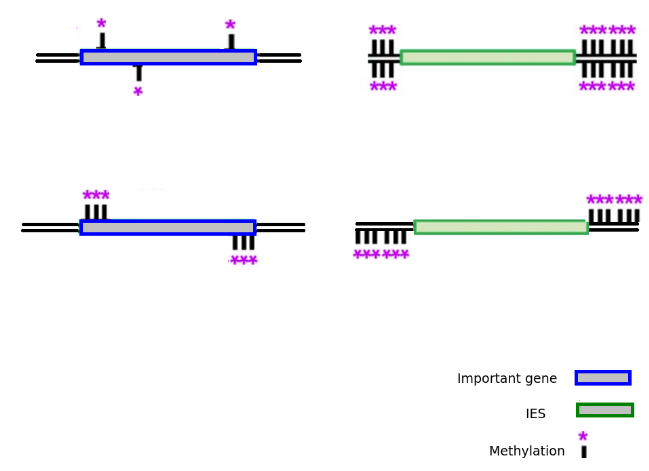
RNA mediated methylation ?
antisens RNA ?
Nucleosome positionning ?
Modified bases: 3 modifications tracked among many others
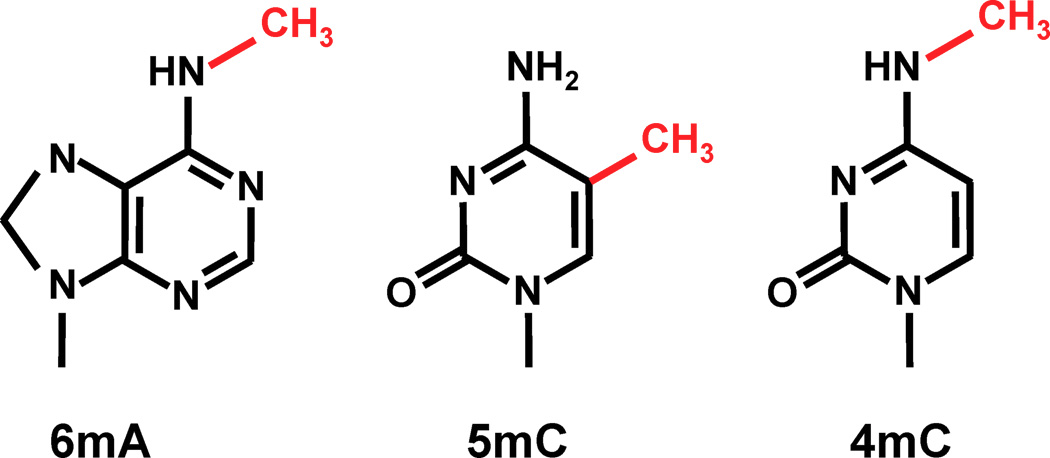
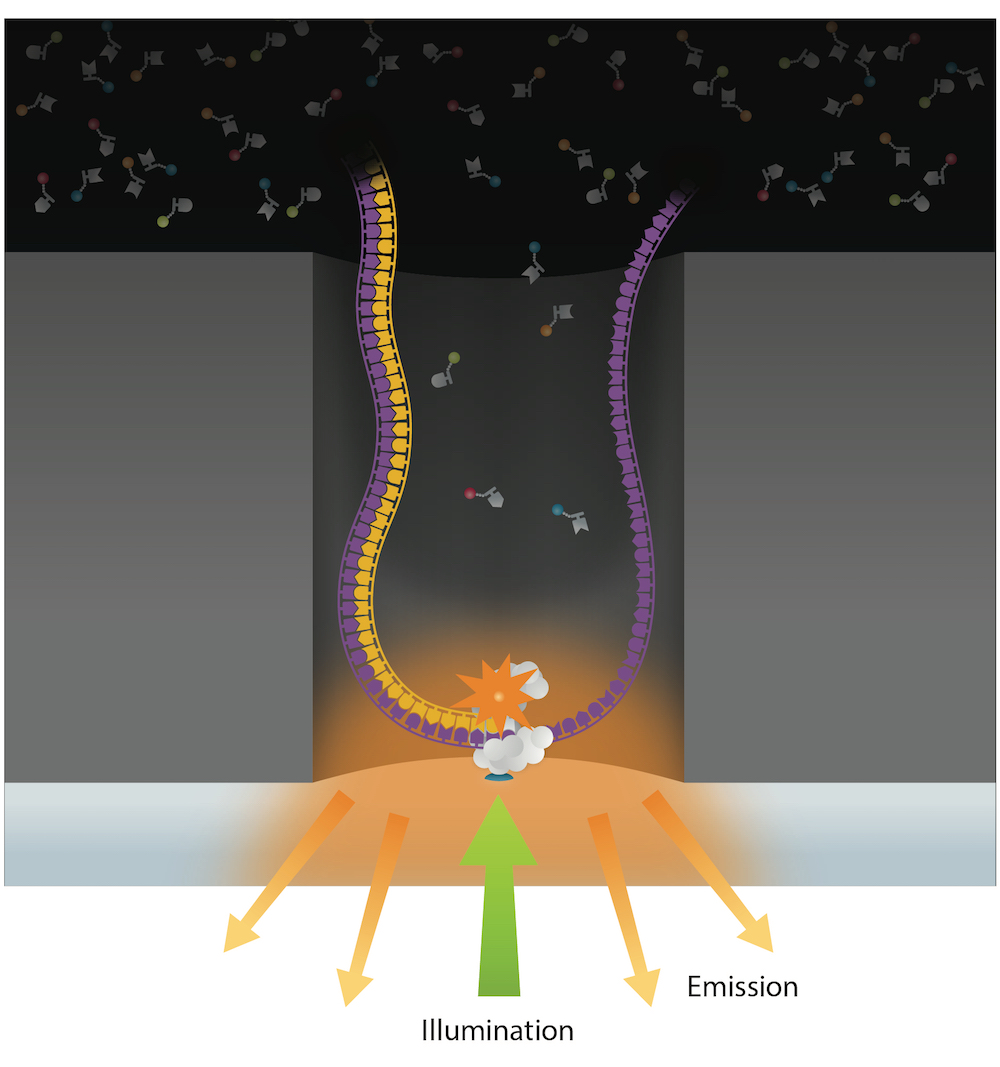
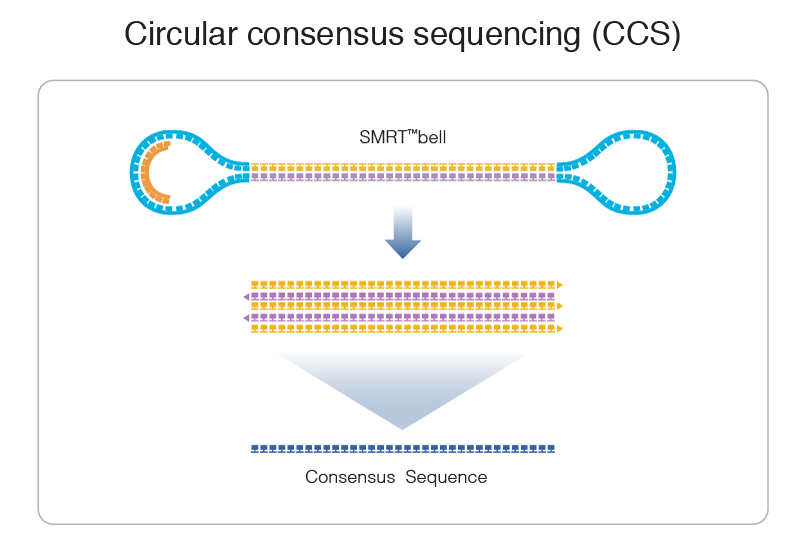
PacBio sequencing
99% accuracy
Max
75% accuracy
Methylation analysis needs local > 25X
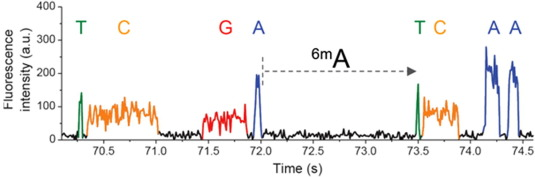
Trained model (ML) allows detection of suspect downturns of polymerase (function of the -3/+8 nt context) --> IPD are captured
Sorting the sequences
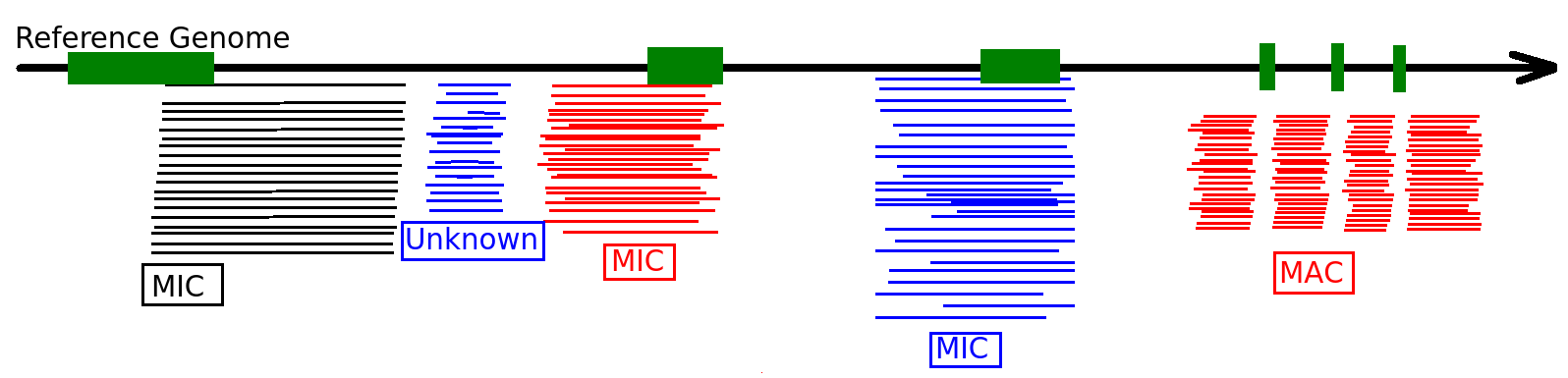
Differential mapping
Sequences that come from MIC
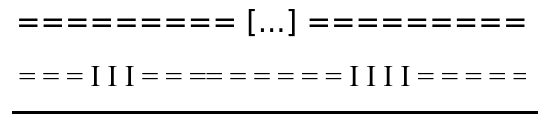
N changes
"=" becomes "I"
Diffential mapping
Sequences that come from MAC
The score threshold
The modification/identification scores are sold by PacBio as PHRED scores
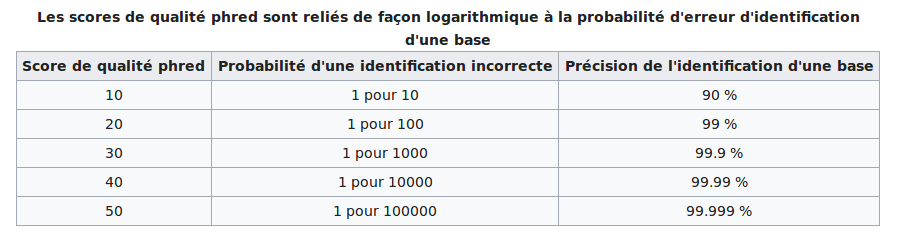
For our experiments: we should have about ~30% of modified bases if this is true
--> Modification scores are NOT prhed scores (at least in our case)
Modification scores overall
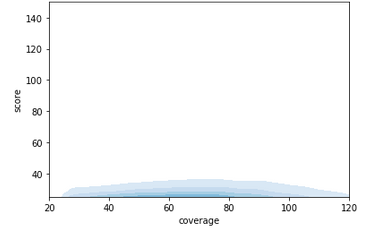
Score distributions: The case of the adenines
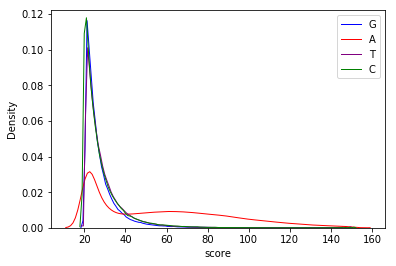
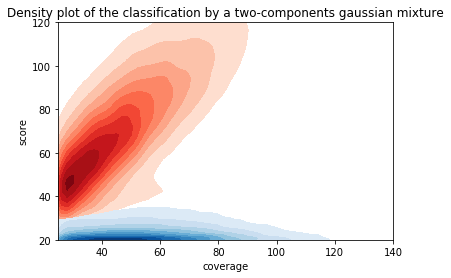
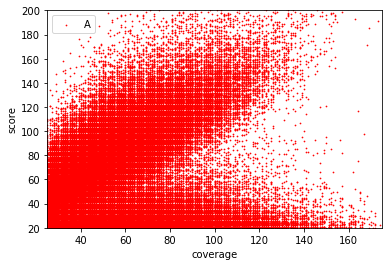
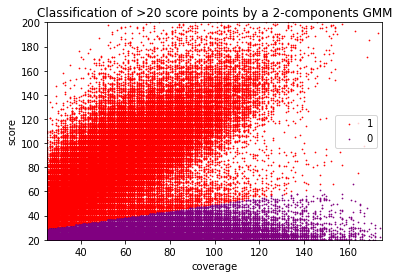
Can we apply the GMM to other samples ?
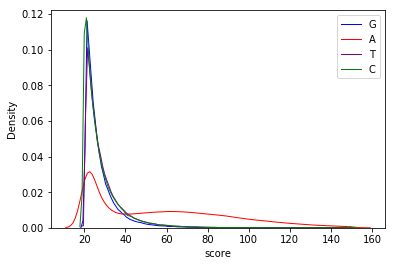
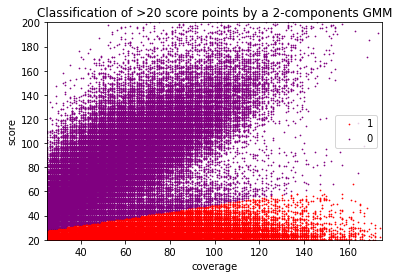
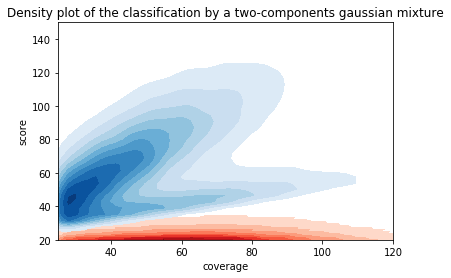
Score distributions: The case of cytosines
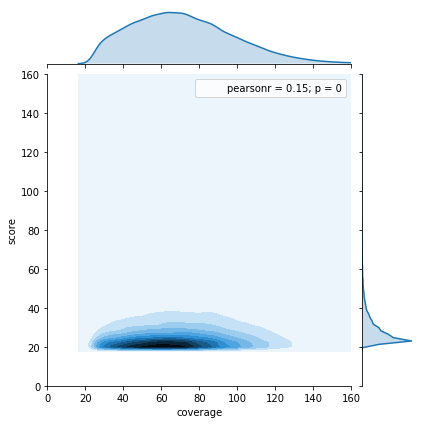
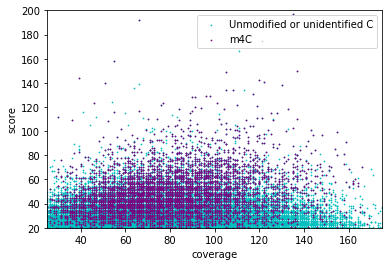
Score distributions: The case of G and T
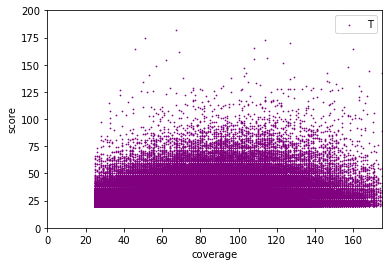
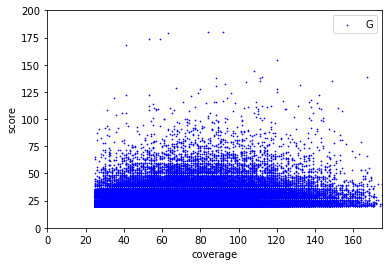
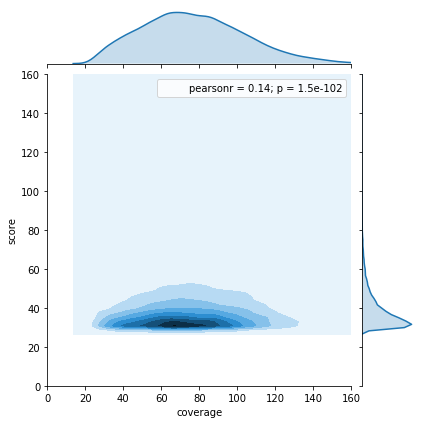
Available Data
HT2
HT6
HTVEG
MAB
MT1A-1B
MT1A-1B-2
MT2
NM9_10
NM4_9_10
WT
Silenced
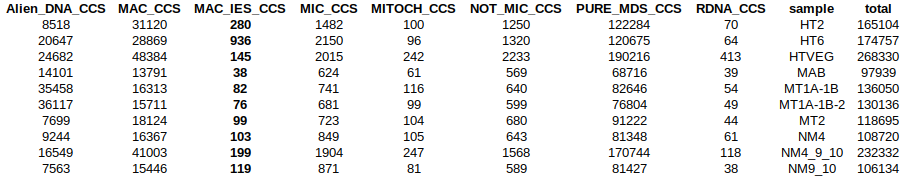
Sorting stats
Using only the GMM
Adenines
All A in GMM
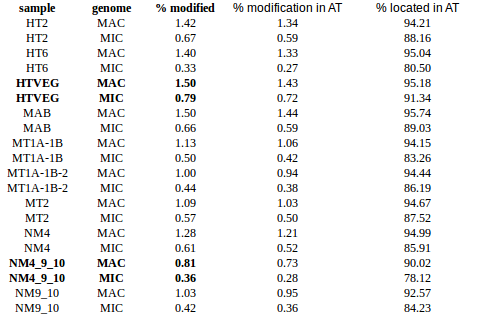
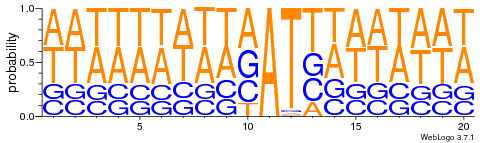
- Same MIC/MAC in AT
- No difference between experiments
- ~95% methylated symetrically
All A in GMM
out the AT
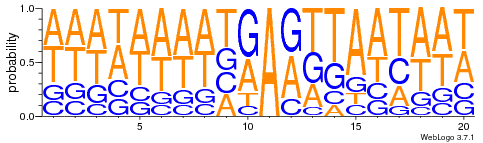
HTVEG
~ Same for every silencing experiment
Lack of sequences (~50 VS ~2.000) don't really allow comparaison between MIC and MAC
Using idQv20 + GMM
Adenines
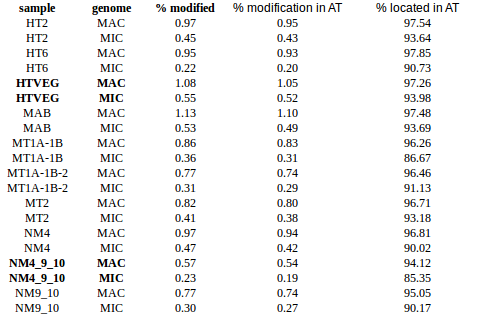
GMM + idQV20
- Raises the % located in AT
- Methylated fraction goes down to 0.9-1%
- NM4-9-10 goes down to 50% in the MAC
- Logos don't change significantly
Cytosines
Qv20/IdQv20

Logo from HTVEG MAC
Identical everywhere
Conclusion
Number of MAC_IES sequences: will it be enough ?
Capping ok
In silico control --> Experimental
Coverage threshold single-hole ?
Threshold is still a +/- open question
TSS to do
ScanRNA dep VS indep
Diminution m6A in some silencings, but which one disappears ?
2.5% m6A in MAC <-> Bad calibration ? MDS missing ? Lack of sensibility ?
NM4_9_10 --> 10% ? Recalibrage par optimisation ?
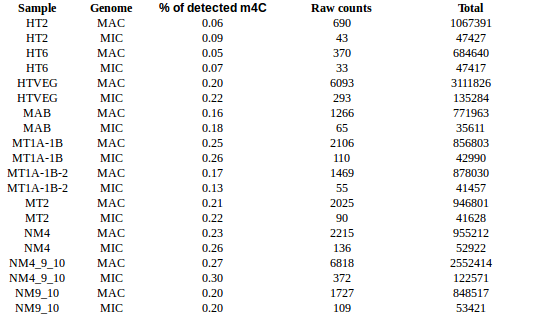
m4C signal: HT2 = HT6 < HTVEG
Other
| N° centre | Centre | Accord | |
|---|---|---|---|
| 207 | Paris Saint-Louis | ok | |
| 230 | Marseille IPC | ok | Faezeh Legrand |
| 233 | Besançon | ok | |
| 234 | Bruxelles St-Luc | ok | |
| 250 | Saint-Etienne | ok | |
| 251 | Caen | ok | |
| 261 | Genève | ok | |
| 262 | la Pitié | ok | |
| 264 | Poitiers | ok | |
| 267 | Bordeaux | ok | |
| 270 | Grenoble | ok | |
| 272 | Tours | ok | |
| 273 | Clermont | ok | |
| 277 | Lille | ok | |
| 301 | Marseille La Timone | ok | |
| 369 | Beyrouth | ok | |
| 523 | Nice | ok | Michaël Loschi |
| 624 | Toulouse | ok | |
| 631 | Paris Robert Debré | ok | |
| 650 | Angers | ok | |
| 659 | Brest | ok | |
| 661 | Rennes pédiatrie | ok | |
| 671 | Lyon CHLS | ok | |
| 671 | Lyon | ok | |
| 672 | Strasbourg | ok | |
| 726 | Liège | ok | |
| 775 | Saint-Antoine | ok | |
| 941 | Rouen adultes | ok | |
| 955 | Amiens | ok | |
| 977 | Limoges | ok | |
| 978 | Bordeaux pédiatrie | ok |

Oila oila
Thx :)
Self and non self
By biocompibens
Self and non self
Lab meeting - 19/06/18
- 118



