Data Analytics, Data Science
& Machine Learning
with R
DISCLAIMER: The images, code snippets...etc presented in this presentation were collected, copied and borrowed from various internet sources, thanks & credit to the creators/owners
(Artificial Intelligence) comprehensively
[ Data Analytics ( internal subfield: overlapping or interrelated ) ]
Agenda
-
What is Data Science?
-
What Data Science Can Do?
-
Prerequisites & Skillset
-
Data Science Workflow
-
Python vs R
-
R Programming
-
Statistics
-
Data Analysis & Visualization
-
Machine Learning
-
Course Curriculum
-
Q & A
What is Data Science?
To gain insights into data through computation, statistics, and visualization to make better decisions
The ability to take data—to be able to understand it, to process it, to extract value from it, to visualize it, to communicate it—that’s going to be a hugely important skill in the next decades - Hal Varian

What Data Science Can Do?
- Predict whether a patient hospitalized due to a heart attach, will have a second heart attach. The prediction is to be based on demographic, diet & clinical measurement for that patient..
- Predict the price of a stock in 6 months from now, on the basis of company performance measures & economic data.
- Identify the risk factors for prostate cancer, based on clinical & demographic variables.
- It can also figure out whether a customer is pregnant or not by capturing their shopping habits from retail stores
- It also knows your age and gender, what brands you like even if you never told., including your list of interests(which you can edit) to decide what kind of ads to show you.
- It can also predict whether or not your relationship is going to last, based on activities and status updates on social networking sites. Police departments in some major cities also know you're going to commit a crime.
- It also tells you what videos you've been watching, what you like to read, & when you're going to quit your job.
- It also guess how intelligent you are how satisfied you are with your life, and whether you are emotionally stable or not -simply based on analysis of the 'likes' you have clicked
this is actually just the tip of the iceberg

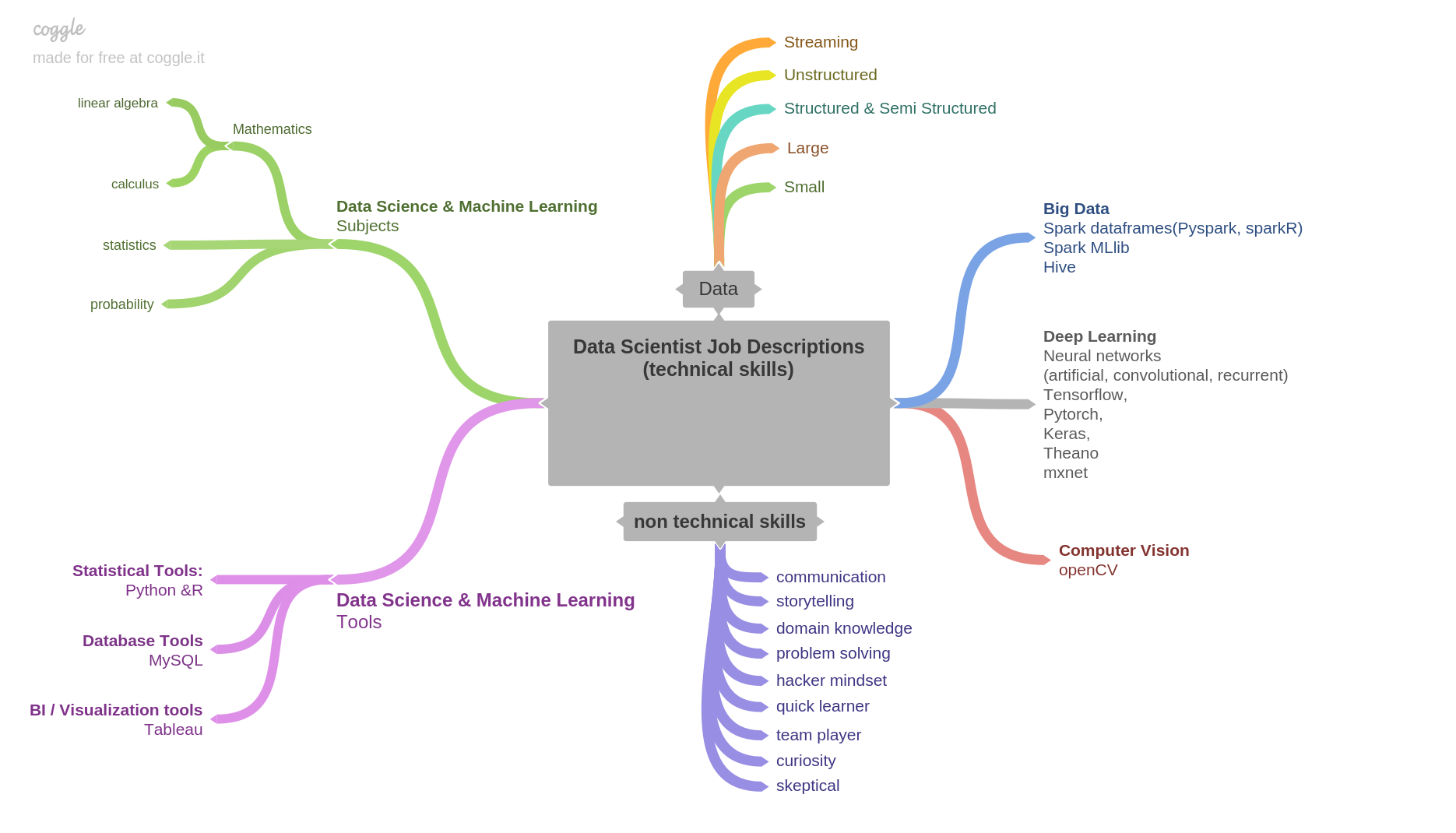
Prerequisites & Skillset



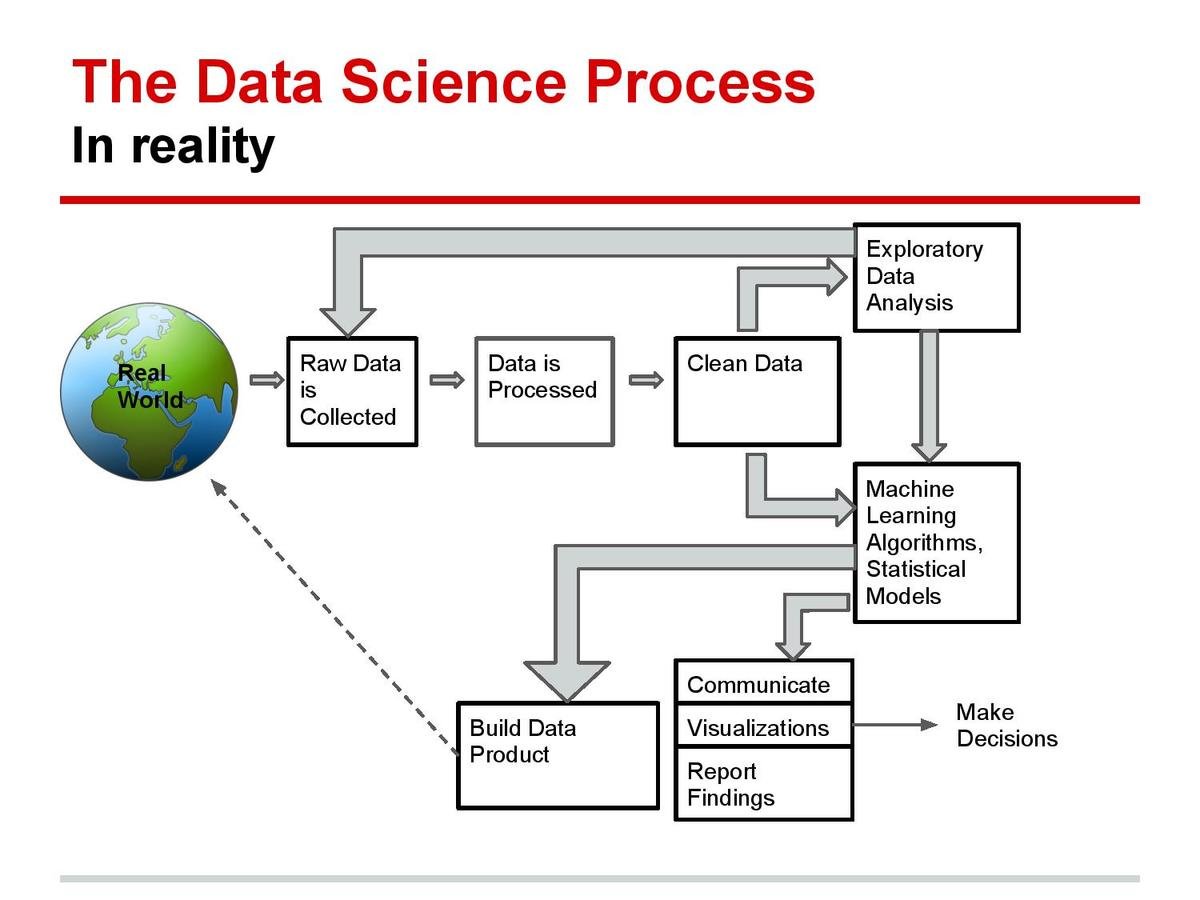
Data Science Workflow
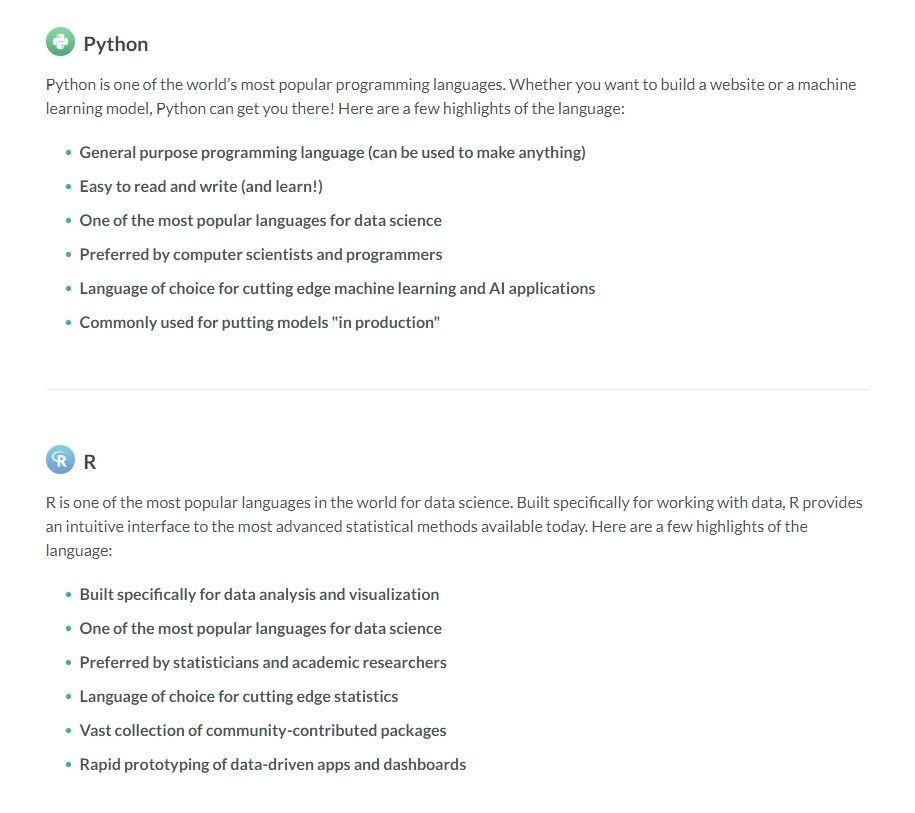
Python vs R
```{R}
# R objects and value storage
myList <- list("Sunday", "Monday", "Wednesday")
print(myList)
myList <- append(myList, "Tuesday", after = 2)
print(myList)
```
```{python}
# Python objects and value storage
x = ["Sunday", "Monday", "Wednesday"]
print(x)
x.append("Tuesday") # append 4 to the list pointed to by x
print(x)
x.insert(2, "Tuesday")
print(x) # y's list is modified as well!
```## ggplot2 - Adv data visualisations
library(ggplot2)
df %>%
filter(gdpPercap<50000) %>%
ggplot(aes(x=log(gdpPercap),y=lifeExp, col=continent, size= pop)) +
geom_point(alpha=0.3) +
geom_smooth(method = lm) +
facet_wrap(~continent)
## machine learning - linear models
model <- lm(df$lifeExp~df$gdpPercap) # univariate simple linear model
summary(model)
model2 <- lm(df$lifeExp~df$gdpPercap+df$pop) # multi variate linear models
summary(model2)R Programming

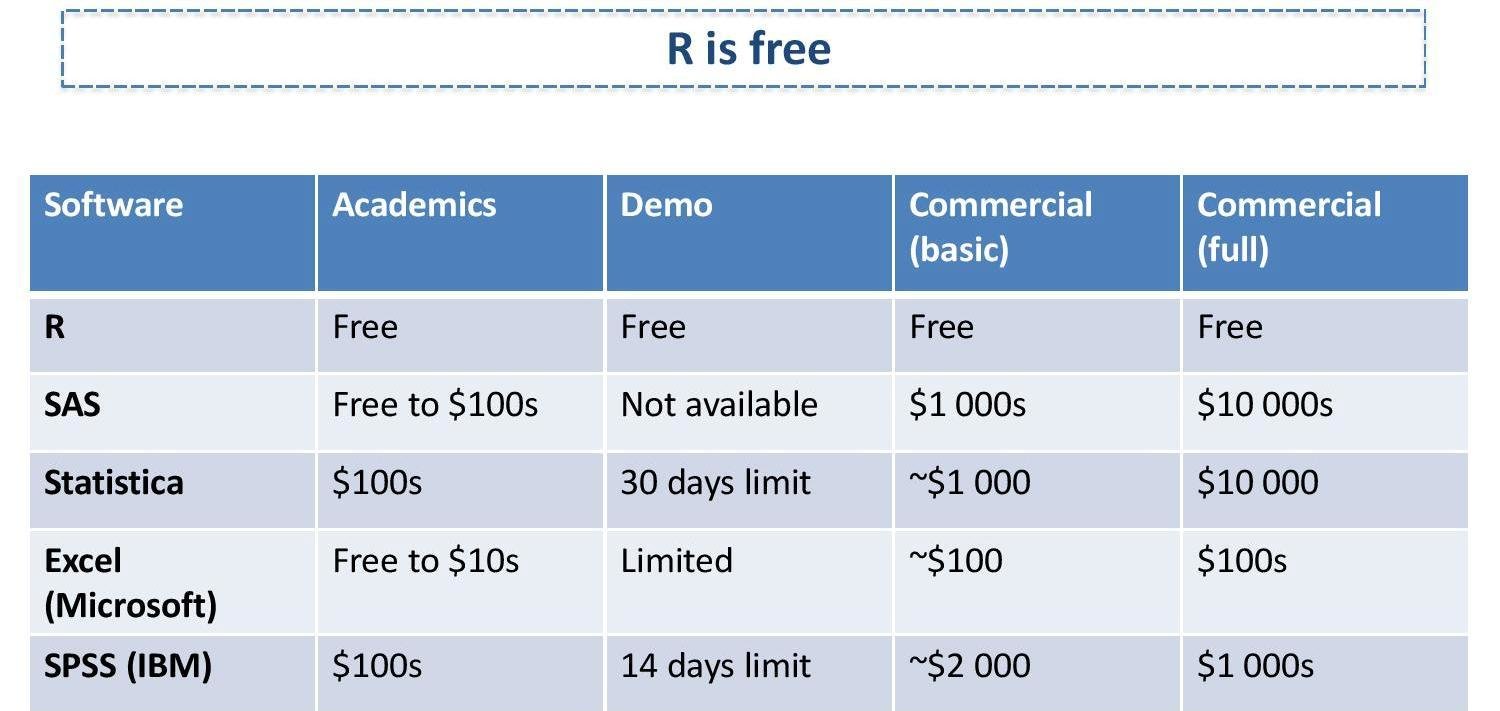
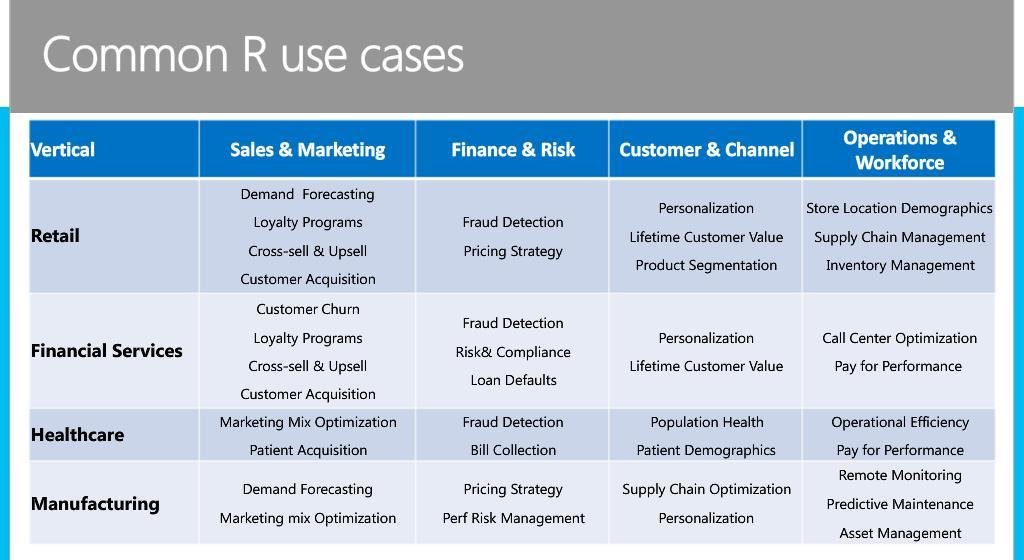
##working with a dataset
install.packages("gapminder") # to install package
library(gapminder) # to load package into session
data("gapminder") # load data to environment
head(gapminder) #sneak peek
?gapminder # dataset details
str(gapminder) # dataset structure
summary(gapminder) # statistical summaries
## understand data using basic visualisations
df <- gapminder
hist(df$lifeExp) # life expectency distribution looks - normal
hist(df$pop) # population distribution looks - skewed
hist(log(df$pop)) # applying log transformation
barplot(table(df$continent)) # continents comparison
boxplot(df$lifeExp~df$continent) # numerical vs categorical
plot(df$lifeExp~df$gdpPercap) # 2 numerical relation between life exp and gdp per cap
plot(df$lifeExp~log(df$gdpPercap)) # log transformation# packages usage
##dplyr - data manipulation
library(dplyr)
df1 <- data.frame(eye_color=rep(c("blue","brown"),c(1,1)),height=round(runif(4, 4.0,
6.5),1),weight=seq(45,60,by= 4)) # creating data frame
print(df1) # looking at data
##applying on data frame
df1 %>%
select(eye_color,height,weight) %>%
filter(eye_color=="blue") %>%
mutate(BMI= weight/(height^2)) %>%
summarise(avg_BMI=mean(BMI))
## applying on gapminder
df %>%
select(country,lifeExp) %>%
filter(country=="India" | country == "Australia") %>%
group_by(country) %>%
summarise(Avg_life=mean(lifeExp))
## applying statistical significance test
df2 <- df %>%
select(country,lifeExp) %>%
filter(country=="India" | country == "Australia")
print(df2)
t.test(data=df2,lifeExp ~ country)Python


Statistics

Elementary statistics

Statistics
Mean, Median, Mode, Standard Deviation, Range, Quartiles, skewness, kurtosis,.. more
#Applying basic statistics in R
data(iris)
class(iris)
mean(iris$Sepal.Length)
sd(iris$Sepal.Length)
var(iris$Sepal.Length)
min(iris$Sepal.Length)
max(iris$Sepal.Length)
median(iris$Sepal.Length)
range(iris$Sepal.Length)
quantile(iris$Sepal.Length)
sapply(iris[1:4], mean, na.rm=TRUE)
summary(iris)
cor(iris[,1:4])
cov(iris[,1:4])
t.test(iris$Petal.Width[iris$Species=="setosa"],
iris$Petal.Width[iris$Species=="versicolor"])
cor.test(iris$Sepal.Length, iris$Sepal.Width)
aggregate(x=iris[,1:4],by=list(iris$Species),FUN=mean)# Applying basic statistics in Python
import pandas as pd
data = {'name': ['Jason', 'Molly', 'Tina', 'Jake', 'Amy'],
'age': [42, 52, 36, 24, 73],
'preTestScore': [4, 24, 31, 2, 3],
'postTestScore': [25, 94, 57, 62, 70]}
df = pd.DataFrame(data, columns = ['name', 'age', 'preTestScore', 'postTestScore'])
print(df)
# Minimum value of preTestScore
df['preTestScore'].min()
# Maximum value of preTestScore
df['preTestScore'].max()
# The sum of all the ages
df['age'].sum()
# Mean preTestScore
df['preTestScore'].mean()
# Median value of preTestScore
df['preTestScore'].median()
#Sample variance of preTestScore values
df['preTestScore'].var()
#Sample standard deviation of preTestScore values
df['preTestScore'].std()
# Cumulative sum of preTestScores, moving from the rows from the top
df['preTestScore'].cumsum()
# Summary statistics on preTestScore
df['preTestScore'].describe()
Data Analysis & Visualization
Data Analysis & Visualizations
Summarize, Scatter plot, Histogram, Box plot, Pie chart, Bar plot, ...... more
# Basic plotting in R
data(iris)
table.iris = table(iris$Species)
table.iris
pie(table.iris)
hist(iris$Sepal.Length)
boxplot(Petal.Width ~ Species, data = iris)
plot(x=iris$Petal.Length, y=iris$Petal.Width, col=iris$Species)
pairs(iris[1:4], main = "Edgar Anderson's Iris Data", pch = 21,
bg = c("red", "green3", "blue")[unclass(iris$Species)])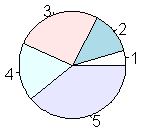
# Basic plotting in Python
import pandas as pd
import matplotlib.pyplot as plt
iris = pd.read_csv("https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data",
header=None)
iris.columns = ['Sepal.Length', 'Sepal.Width', 'Petal.Length', 'Petal.Width',
'Species']
iris.head()
iris['Sepal.Length'].plot(kind='hist')
iris.plot(kind='box')
plt.show()
iris.plot(kind='scatter', x='Sepal.Length', y='Sepal.Width')
plt.show()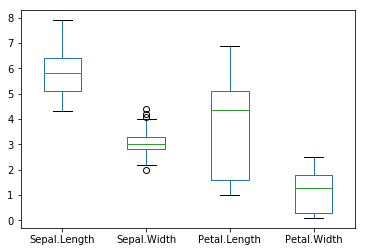
#console and script
> x = 7
> x + 9
[1] 16
#comments
# COMMENTS ARE SUPER IMPORTANT so we learned about them
#graphics
x = rnorm(1000, mean = 100, sd = 3)
hist(x)
#getting help
# if you know the function name, but not how to use it:
?chisq.test
# if you know what you want to do, but don't know the function name:
??chisquare
#data types
# character vector
> y = c("apple", "apple", "banana", "kiwi", "bear", "strawberry", "strawberry")
> length(y)
[1] 7
# numeric vector
> numbers = rep(3, 99)
> numbers
[1] 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
[39] 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
[77] 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
#matrices
> mymatrix = matrix(c(10, 15, 3, 29), nrow = 2, byrow = TRUE)
> mymatrix
[,1] [,2]
[1,] 10 15
[2,] 3 29
> t(mymatrix)
[,1] [,2]
[1,] 10 3
[2,] 15 29
> solve(mymatrix)
[,1] [,2]
[1,] 0.1183673 -0.06122449
[2,] -0.0122449 0.04081633
> mymatrix %*% solve(mymatrix)
[,1] [,2]
[1,] 1 0
[2,] 0 1
> chisq.test(mymatrix)
Pearson's Chi-squared test with Yates' continuity correction
data: mymatrix
X-squared = 5.8385, df = 1, p-value = 0.01568
#data frames (the mother of all R data types)
# set working directory
setwd("~/Documents/R_intro")
# read in a dataset
wages = read.table("wages.csv", sep = ",", header = TRUE)
#exploratory data analysis
> names(wages)
[1] "edlevel" "south" "sex" "workyr" "union" "wage" "age"
[8] "race" "marital"
> class(wages$marital)
[1] "integer"
> table(wages$union)
not union member union member
438 96
> summary(wages$workyr)
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.00 8.00 15.00 17.82 26.00 55.00
> nrow(wages)
[1] 534
> length(which(is.na(wages$sex)))
[1] 0
> linmod = lm(workyr ~ age, data = wages)
> summary(linmod)
hist(wages$wage, xlab = "hourly wage", main = "wages in our dataset", col = "purple")
plot(wages$age, wages$workyr, xlab = "age", ylab="years worked", main = "age vs. years worked")
abline(lm(wages$workyr ~ wages$age), col="red", lwd = 2)
mtcars[mtcars$mpg>=20,c('gear','mpg')]
aggregate(. ~ gear,mtcars[mtcars$mpg>=20,c('gear','mpg')],mean)
subset(aggregate(. ~ gear,mtcars[mtcars$mpg>=20,c('gear','mpg')],mean),mpg>25)
#output
gear mpg
2 4 25.74
3 5 28.20Machine Learning


myData <- data.frame(age=c(10,20,30), weight=c(100,200,300))
myPersonsAge <- data.frame(age=c(20,25,30))
myTrainModel <- lm(weight~age,data = myData)
predict(myTrainModel,myPersonsAge)Machine Learning Intro
# install.packages("kernlab")
# install.packages("caret")
library(kernlab);library(caret)
data("spam")
predictors <- names(churnTrain)[names(churnTrain)=='churn']
predictors
set.seed(1)
inTrain <- createDataPartition(y=spam$type, p=0.75, list = FALSE)
training <- spam[inTrain,]
testing <- spam[-inTrain,]
dim(training)
set.seed(32343)
modelFit <- train(type~.,data = training, method='glm')
modelFit
modelFit$finalModel
predictions <- predict(modelFit,newdata=testing)
predictions
confusionMatrix(predictions, testing$type)
Generalized Linear Models (Logistic regression)
Machine Learning Algorithms
Linear regression
Decision trees
K-means
# Load required packages
library(ggplot2)
library(datasets)
# Load data
data(iris)
# Set seed to make results reproducible
set.seed(20)
# Implement k-means with 3 clusters
iris_cl <- kmeans(iris[, 3:4], 3, nstart = 20)
iris_cl$cluster <- as.factor(iris_cl$cluster)
# Plot points colored by predicted cluster
ggplot(iris, aes(Petal.Length, Petal.Width, color = iris_cl$cluster)) + geom_point()# Include required packages
library(party)
library(partykit)
# Have a look at the first ten observations of the dataset
print(head(readingSkills))
input.dat <- readingSkills[c(1:105),]
# Grow the decision tree
output.tree <- ctree(
nativeSpeaker ~ age + shoeSize + score,
data = input.dat)
# Plot the results
plot(as.simpleparty(output.tree))# Load required packages
library(ggplot2)
# Load iris dataset
data(iris)
# Have a look at the first 10 observations of the dataset
head(iris)
# Fit the regression line
fitted_model <- lm(Sepal.Length ~ Petal.Width + Petal.Length, data = iris)
# Get details about the parameters of the selected model
summary(fitted_model)
# Plot the data points along with the regression line
ggplot(iris, aes(x = Petal.Width, y = Petal.Length, color = Species)) +
geom_point(alpha = 6/10) +
stat_smooth(method = "lm", fill="blue", colour="grey50", size=0.5, alpha = 0.1)Support vector machine
#svm
library(e1071)
# quick look at the data
plot(iris)
# feature importance
plot(iris$Sepal.Length, iris$Sepal.Width, col=iris$Species)
plot(iris$Petal.Length, iris$Petal.Width, col=iris$Species)
#split data
s <- sample(150, 100)
col <- c('Petal.Length','Petal.Width','Species')
iris_train <- iris[s,col]
iris_test <- iris[-s,col]
#create model
svmfit <- svm(Species ~ ., data = iris_train, kernel="linear", cost=.1, scale = FALSE)
print(svmfit)
plot(svmfit, iris_train[, col])
tuned <- tune(svm, Species~., data = iris_train, kernel='linear', ranges = list(cost=c(0.001, 0.01,.1, 1.10, 100)))
summary(tuned)
p <- predict(svmfit, iris_test[,col], type='class')
plot(p)
table(p, iris_test[,3])
mean(p==iris_test[,3])
Machine Learning Algorithms
k-nearest neighbours(knn) classifier
# importing necessary libraries
from sklearn import datasets
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import train_test_split
# loading the iris dataset
iris = datasets.load_iris()
# X -> features, y -> label
X = iris.data
y = iris.target
# dividing X, y into train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state = 0)
# training a KNN classifier
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors = 7).fit(X_train, y_train)
# accuracy on X_test
accuracy = knn.score(X_test, y_test)
print (accuracy)
# creating a confusion matrix
knn_predictions = knn.predict(X_test)
cm = confusion_matrix(y_test, knn_predictions) 
Data Science Skills are in High Demand Across Industries
(Linkedin Report & Payscale)
Course Curriculum


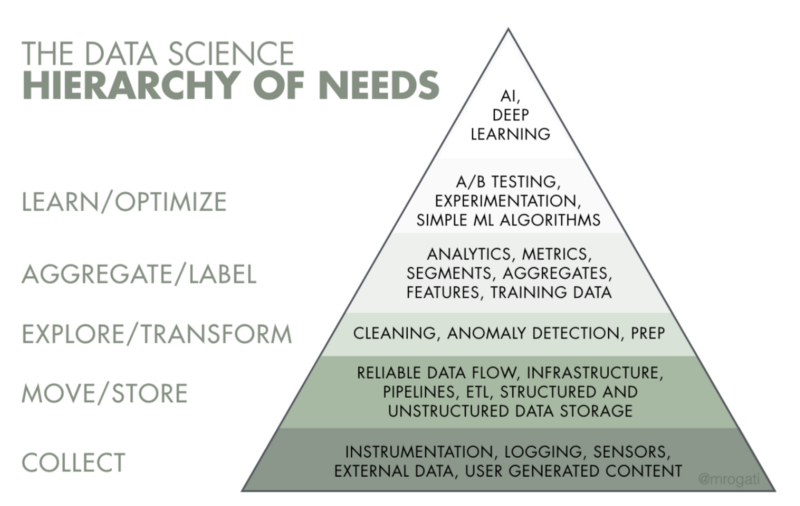
The AI Hierarchy
Difference between Artificial intelligence(or AI), Machine Learning and Deep Learning?

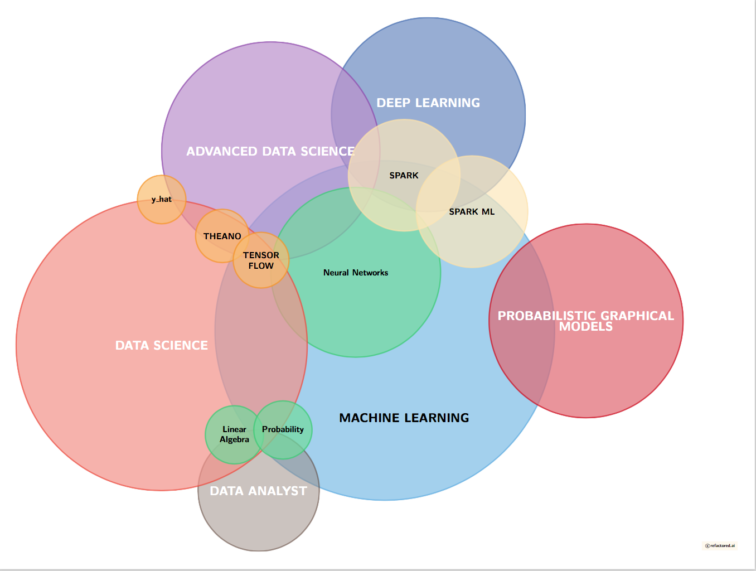
Overlapping Skillsets
Overlapping Skillsets

Data Science & Machine Learning with Python and R - demo
By Data Science Portal
Data Science & Machine Learning with Python and R - demo
Data Science & Machine Learning with Python and R
- 82



