


\text{Embedding-based Instance Segmentation in Microscopy}
\text{\textbf{Manan Lalit}, Pavel Tomancak, Florian Jug}
\text{Publication}
\text{Project Page}


\text{Early development of an organism}
\text{Nuclei}
\text{Membrane}
\text{Light sheet Imaging}
\text{Time-lapse recording of Embryo 1}
\text{Confocal Imaging}
\text{Nuclei}
\text{Gene}
\text{Membrane}
\text{Associate Nuclei based on \textbf{Morphology}}
\text{Instance Segmentation}
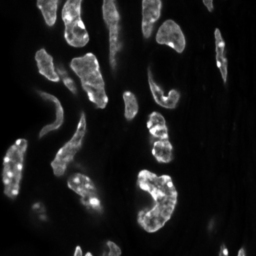
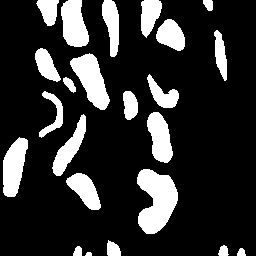



\text{Unique Segmentation of Each Nucleus/Cell}
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
\text{Instance Segmentation}
\text{Desirable properties of Instance Segmentation Approaches}
\text{}
\text{non-parametric object representation}

\text{directly optimize over Intersection over union (IoU) metric during training}

\text{low GPU memory requirement}


\text{Embedding-based approaches}
\text{Could we predict a tag or embedding for each pixel such that:}
\text{pixels belonging to same object are clustered together in the embedding space, and}
\text{pixels belonging to different objects are clustered away from each other in }
\text{the embedding space?}
\text{and Clustering Bandwidth", Neven et al, 2019}
\text{``Instance Segmentation by Jointly Optimizing Spatial Embeddings }
\text{.}
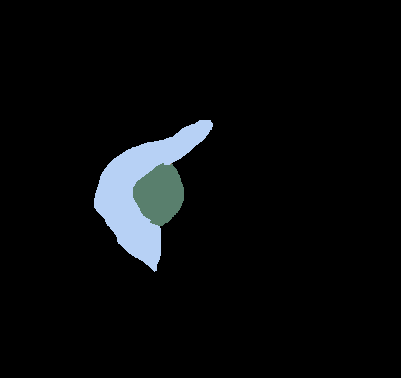
\text{Choice of GT center}
\text{Choice of GT center}

\text{.}
\text{Choice of GT center}

\text{.}

\text{center = centroid}
\text{predicted instance at inference}
\text{Choice of GT center}

\text{.}
\text{center = medoid}
\text{predicted instance at inference}


\text{Preparing new 3d datasets}




\text{Live Embryo}
\textit{in-situ} \text{ specimen}
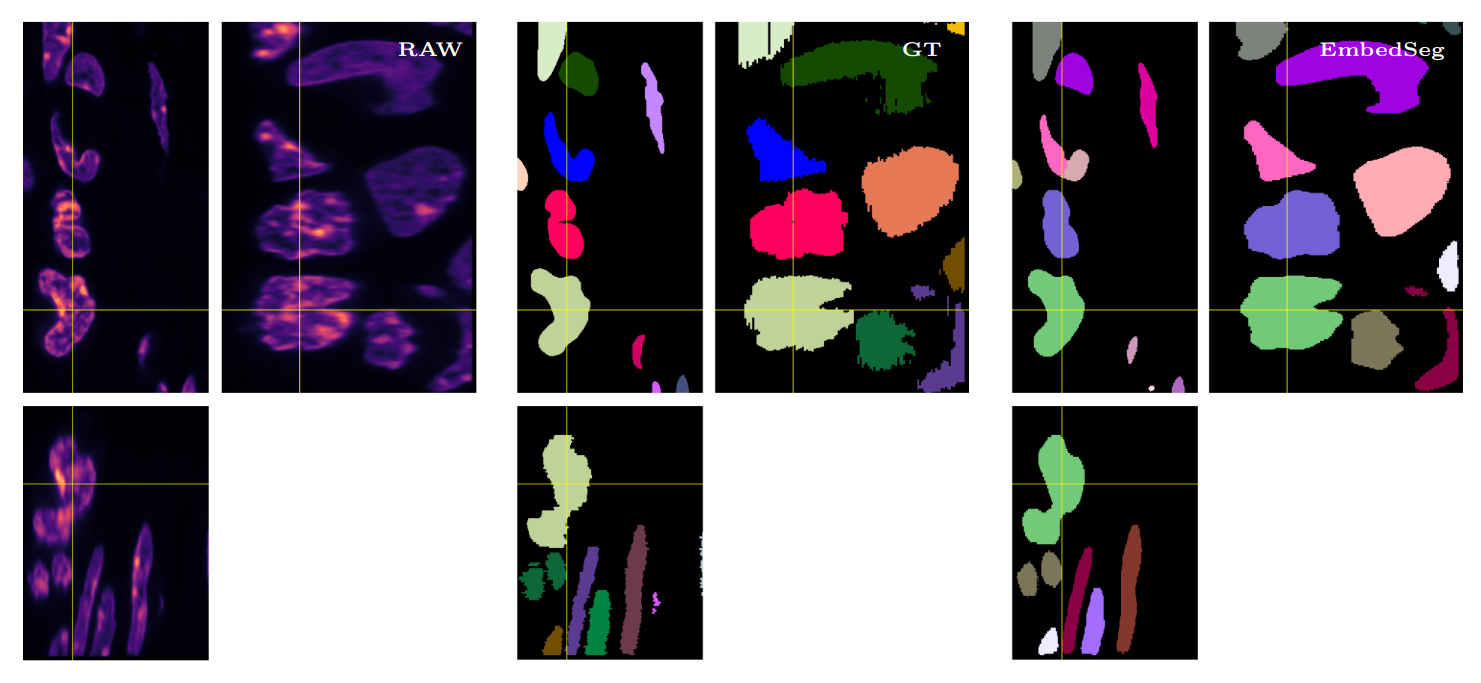
\text{Qualitative Results}
\text{Our Contributions}
\text{Four, new 3d datasets and annotations are made publicly available}

\text{All code and example notebooks are provided}

\text{Demonstrated success of spatial-embedding approaches on biomedical data}

\text{Extended architecture and method to 3d}

\text{Showed that medoid center embedding \& test-time augmentation further boost results}

\text{Four, new 3d datasets and annotations are made publicly available}

\text{All code and example notebooks are provided}

\text{Demonstrated success of spatial-embedding approaches on biomedical data}

\text{Extended architecture and method to 3d}

\text{Showed that medoid center embedding \& test-time augmentation further boost results}

\text{https://github.com/juglab/EmbedSeg}

\text{Our Contributions}
\text{Four, new 3d datasets and annotations are made publicly available}

\text{All code and example notebooks are provided}

\text{Demonstrated success of spatial-embedding approaches on biomedical data}

\text{Extended architecture and method to 3d}

\text{Showed that medoid center embedding \& test-time augmentation further boost results}


\text{Provided a light-weight GPU-friendly code which benefits from virtual batching}
\text{Our Contributions}
\text{Four, new 3d datasets and annotations are made publicly available}

\text{All code and example notebooks are provided}

\text{Demonstrated success of spatial-embedding approaches on biomedical data}

\text{Extended architecture and method to 3d}

\text{Showed that medoid center embedding \& test-time augmentation further boost results}

\text{https://github.com/juglab/EmbedSeg-napari}
\text{Provided a light-weight GPU-friendly code which benefits from virtual batching}

\text{Our Contributions}

















\text{Acknowledgements}
\text{Thank you for listening!}
\newline
\text{Any questions?}
\text{Alex Krull}
\text{Matthias Arzt}
\text{Tim-Oliver Buchholz}
\text{Mangal Prakash}
\text{Anna Goncharova}
\text{Nuno Martins}
\text{Tobias Pietzsch}
\text{Deborah Schmidt}
\text{Florian Jug}
\text{Pavel Tomancak}
\text{Joran Deschamps}
\text{Marina Cuenca}
\text{Giulia Serafini}
\text{Yu-Wen Hsieh}
\text{Bruno Vellutini}
\text{Johannes Girstmair}
\text{Mette Thorsager}

\text{Tom Burke}

\text{Pavel Mejstrik}

\text{Anais Bailles}


MIDL_2021_Oral
By Manan Lalit
MIDL_2021_Oral
MIDL, 2021 (Long Oral)
- 27


