STIMULUS DETECTION USING NEUROIMAGING
INTRODUCTION
Statistical machine learning methods are increasingly used for
neuroimaging data analysis.
Their main virtue is their ability to model
high-dimensional datasets, e.g., multivariate analysis of activation
images or resting-state time series.
By considering different functional neuroimaging applications, we illustrate how scikit-learn, a Python machine learning library, can be used to perform some key analysis steps.
AIM & OBJECTIVE
Interest in applying statistical machine learning to neuro-imaging data analysis is growing. Neuroscientists use it as a powerful, albeit complex, tool for statistical inference. The tools are developed by computer scientists who may lack a deep understanding of the neuroscience questions.
This poject aims to fill the gap between machine learning and neuro-imaging by demonstrating how a general-purpose machine-learning toolbox, scikit-learn, can provide state-of-the-art methods for neuro-imaging analysis while keeping the code simple and understandable by both worlds.
DECODING
Decoding refers to learning a model that predicts behavioral or phenotypic variables from brain imaging data.
-> Classification Problem
-> Experiment in "Haxby et al. (2001)"
:- Visual Stimuli from 8 different categories; 6 Subjects; 12 Sessions;
-> Experiment
:- Visual Stimuli from 2 different categories; 1 Subjects;
-> Univariate feature selection and Support Vector Machines (SVM)
THE MENTAL REPRESENTATION OF OBJECTS IN THE BRAIN.
Feature Selection
Scikit-learn offers a panel of strategies to select features. In supervised learning, the most popular feature selection method is the F-test.

feature_selection = SelectKBest(f_classif,
k=500)
clf = SVC(kernel=’linear’)
X_reduced = feature_selection.fit_
transform(X)
clf.fit(X_reduced, y)
### Look at the discriminating weights
coef = clf.coef_
### Reverse feature selection
coef = feature_selection.inverse_transform
(coef)One can choose to take a percentile of the features (SelectPercentile), or a fixed number of features (SelectKBest)
ENCODING
Encoding refers to predicting the imaging data given external variables, such as stimuli descriptors.
An encoding approach allows one to quantify for each voxel to what extent its variability is captured by the model. A popular evaluation method is the predictive R2 score, which uses a prediction on left out data to quantify the decrease in residual norm brought about by fitting a regression function as opposed to fitting a constant.
BRAIN ACTIVITY AND DECODING IMAGES
RESTING-STATE AND FUNCTIONAL CONNECTIVITY ANALYSIS
Even in the absence of external behavioral or clinical variable, studying the structure of brain signals can reveal interesting information as brain activation exhibits coherent spatial patterns during rest.
Resting state fMRI is unlabeled data in the sense that the brain activity at a given instant in time cannot be related to an output variable. In machine learning, this class of problems is known as unsupervised learning.
TECHNOLOGIES USED

Python is a general-purpose, interpreted, high-level programming language. Python allows programmers to use different programming styles to create simple or complex programs, get quicker results and write code almost as
if speaking in a human language.
HTML
PYTHON
HTML is a computer language devised to allow website creation. These websites can then be viewed by anyone else connected to the Internet. It is relatively easy to learn, with the basics being accessible to most people in one sitting; and quite powerful in what it allows you to create.
Cascading Style Sheets (CSS) is a style sheet language used for describing the presentation of a document written in a markup language. Although most often used to set the visual style of web pages and user interfaces written in HTML and XHTML, the language can be applied to any XML document, including plain XML, SVG and XUL, and is applicable to rendering in speech, or on other media.
BOOTSTRAP
CSS
Bootstrap is a free and open-source front-end framework for developing websites and web applications. It contains HTML- and CSS-based design templates for typography, forms, buttons, navigation and other interface components, as well asoptional JavaScript extensions.
PYTHON
LIBRARIES
USED
It is a library for the Python programming language, adding support for large, multi-dimensional arrays and matrices, along with a large collection of high-level mathematical functions to operate on these arrays.
SCIPY
NUMPY
It contains modules for optimization, linear algebra, integration, interpolation, special functions, FFT, signal and image processing, ODE solvers and other tasks common in science and engineering.
It is a plotting library for the Python programming language and its numerical mathematics extension NumPy.
NIBABEL
MATPLOTLIB
This package provides read +/- write access to some common medical and neuroimaging file formats.
SCIKIT-LEARN AND THE MACHINE LEARNING ECOSYSTEM
Scikit-learn is a general purpose machine learning library written in Python. It provides efficient implementations of state-of-the-art algorithms, accessible to non-machine learning experts, and reusable across scientific disciplines and application fields.
In scikit-learn, all objects and algorithms accept input data in the
form of 2-dimensional arrays of size samples × features. Scikit-learn objects share a uniform set of methods that depends on their purpose:
- estimators can fit models from data,
- predictors can make predictions on new data
- transformers convert data from one representation to another.
DATA PREPARATION
- SPATIAL RESAMPLING :- Neuroimaging data often come as Nifti files, 4-dimensional data along with a transformation matrix used to compute voxel locations from array indices to world coordinates.
- SIGNAL CLEANING :- Due to its complex and indirect acquisition process, neuroimaging data often have a low signal-to-noise ratio. They contain trends and artifacts that must be removed to ensure maximum machine learning algorithms efficiency.
- MASKING :-Neuroimaging data are represented in 4 dimensions: 3 spatial dimensions, and one dimension to index time or trials. Scikit-learn algorithms, on the other hand, only accept 2-dimensional samples × features matrices.Applying the mask is made easy by NumPy advanced index-ing using boolean arrays.
- DATA VISUALIZATION :- It is the graphic representation of data. It involves producing images that communicate relationships among the represented data to viewers of the images. This communication is achieved through the use of a systematic mapping between graphic marks and data values in the creation of the visualization.
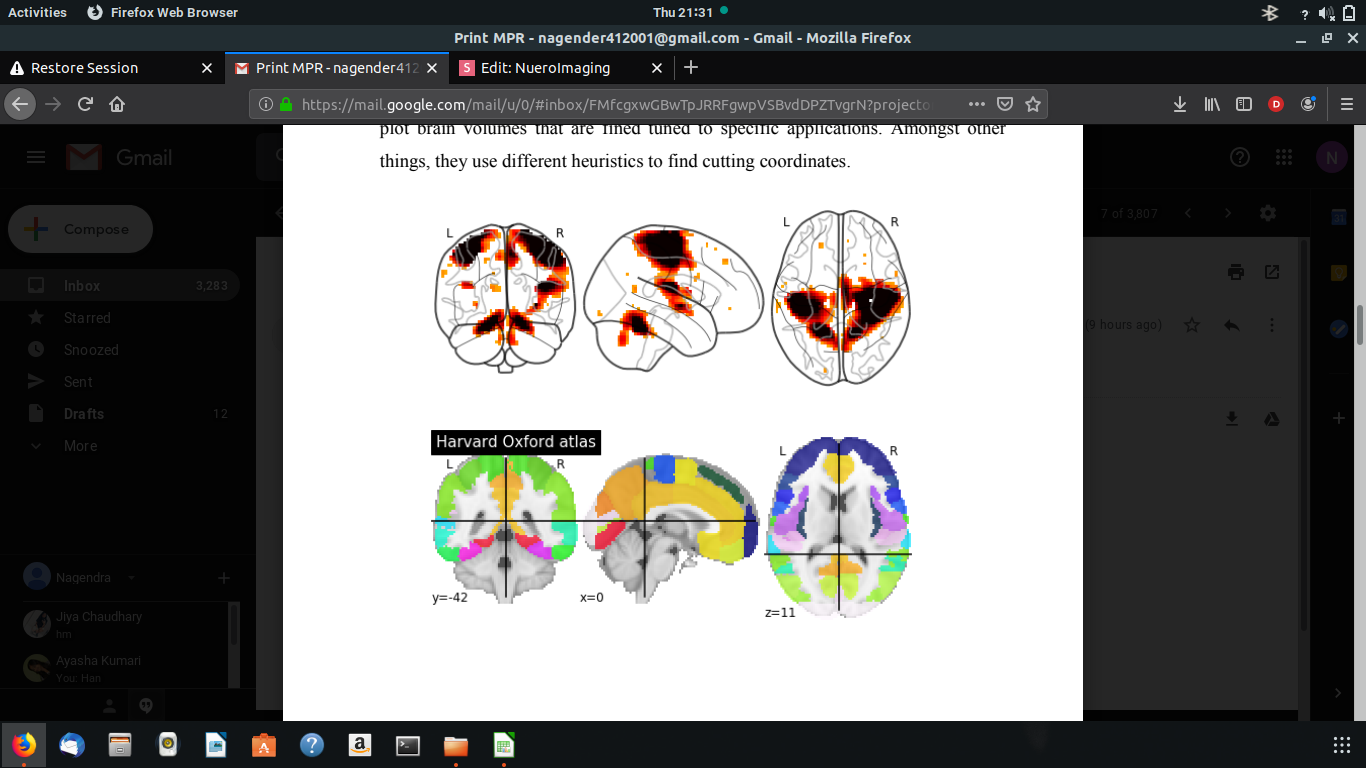
VISUALISATION OF NIFTI FILES
NIfTI stands for Neuro-imaging Information Technology Initiative. It's a standard representation of images, and it's the most common used type of analytic file. Nilearn comes with plotting function to display brain maps coming from Nifti-like images, in the nilearn plotting module. Nilearn has a set of plotting functions to plot brain volumes that are fined tuned to specific applications. Among other things, they use different heuristics to find cutting coordinates.
MIYAWAKI EXPERIMENT
Perceptual experience consists of an enormous number of possible states. Previous fMRI studies have predicted a perceptual state by classifying brain activity into prespecified categories.Constraint-free visual image reconstruction is more challenging, as it is impractical to specify brain activity for all possible images.
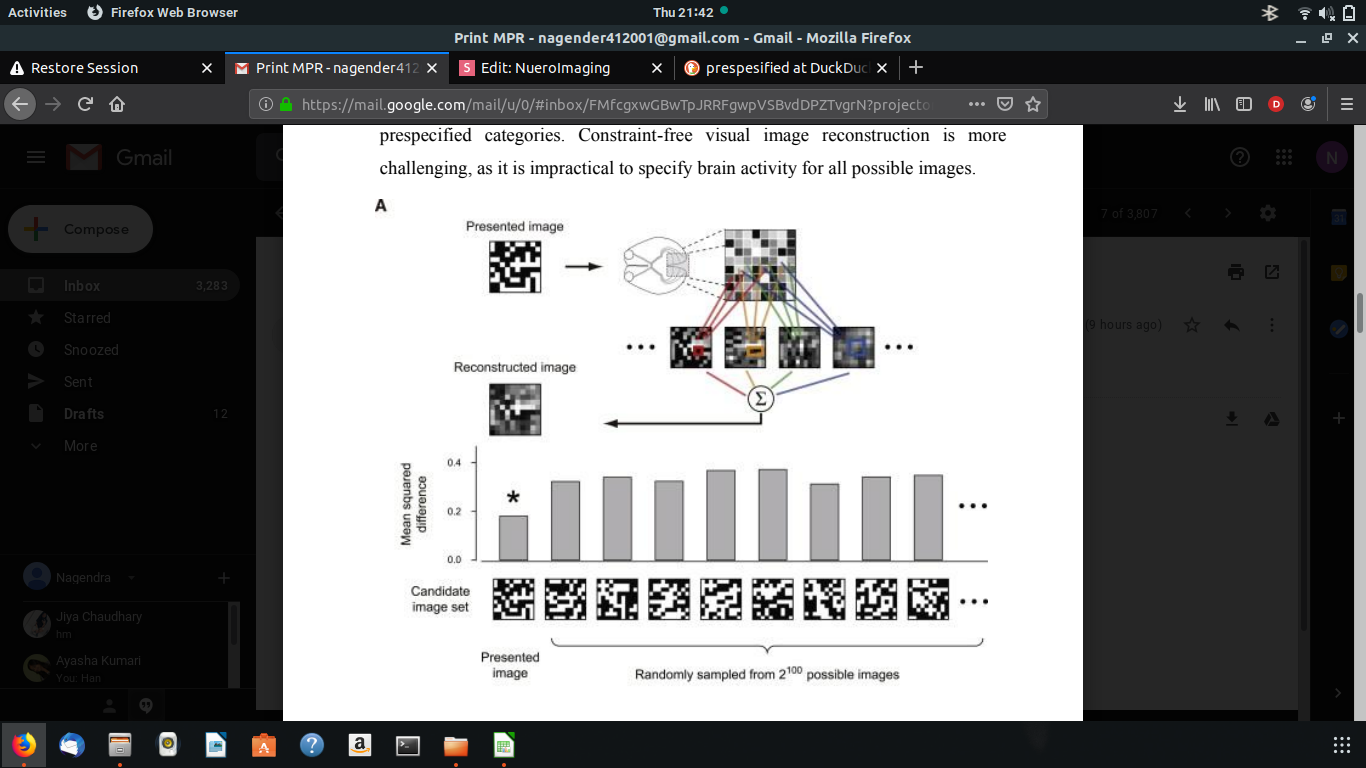
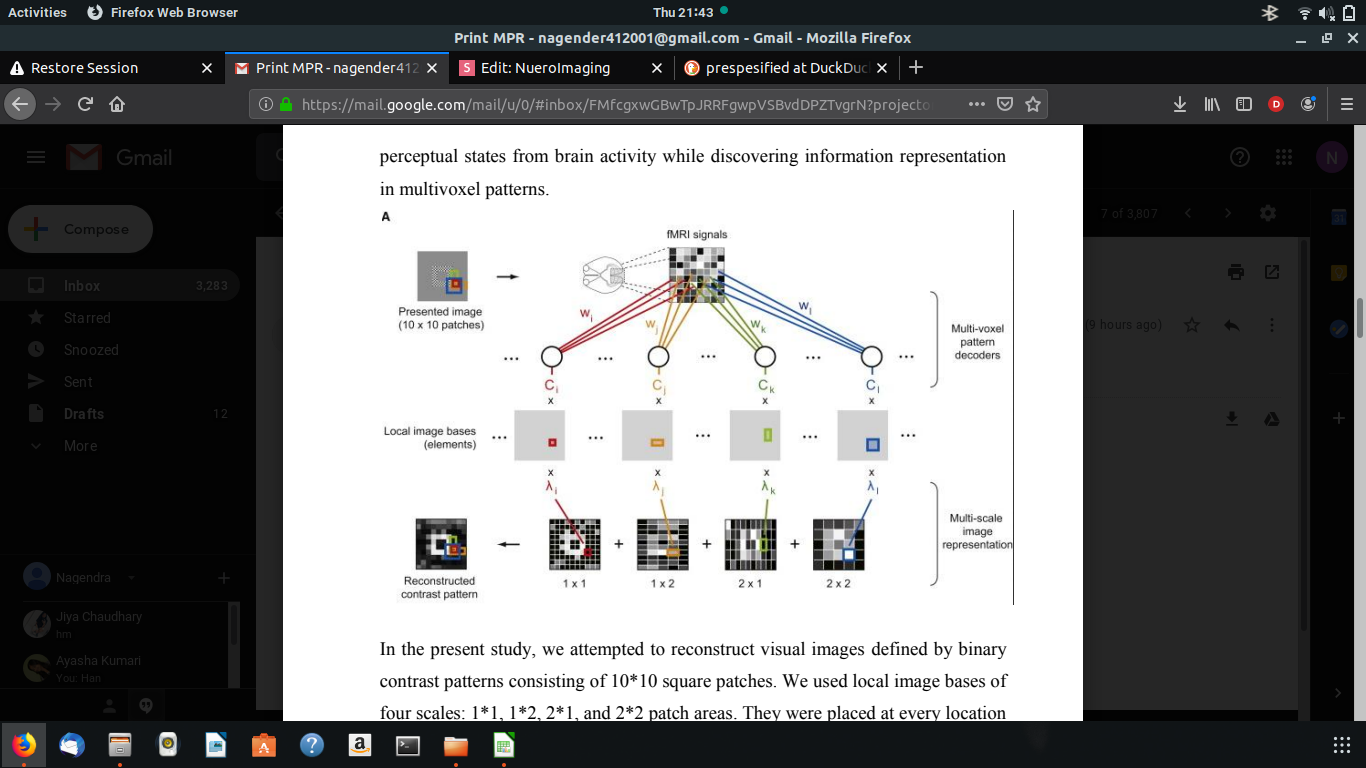
In the present study, we attempted to reconstruct visual images defined by binary contrast patterns consisting of 10*10 square patches. We used local image bases of four scales: 1*1, 1*2, 2*1, and 2*2 patch areas. They were placed at every location in the image with overlaps. Although image elements larger than 2*2 or those with non-rectangular shapes could be used, the addition of such elements did not mprove the reconstruction performance. fMRI signals were measured while subjects viewed a sequence of visual images consisting of binary contrast patches on a 10*10 grid. In the ‘‘random image session,’’ a random pattern was presented for 6s followed by a 6s rest period. A total of 440 different random images were shown (each presented once).
FUNCTIONAL LOCALIZER DATASET
The Functional Localizer is a simple and fast acquisition procedure based on a 5-minute functional magnetic resonance imaging (fMRI) sequence that can be run as easily and as systematically as an anatomical scan. This protocol captures the cerebral bases of auditory and visual perception, motor actions, reading, language comprehension and mental calculation at an individual level. Individual functional maps are reliable and quite precise.
ANALYSIS OF MOTOR TASK FROM LOCALIZER DATASET
This example shows the results obtained in a massively univariate analysis performed at the inter-subject level with various methods. We use the [left button press (auditory cue)] task from the Localizer dataset and seek association between the contrast values and a variate that measures the speed of pseudo-word reading. No confounding variate is included in the model.
- A standard Anova is performed. Data smoothed at 5 voxels FWHM are used.
- A permuted Ordinary Least Squares algorithm is run at each voxel. Data smoothed at 5 voxels FWHM are used.
HAXBY DATASET
It is a block design fMRI dataset from a study on face and object representation in human ventral temporal cortex. It consists of 6 subjects/persons with 12 sessions per subject. In each run, the subjects passively viewed greyscale images of eight object categories, grouped in 24s blocks separated by rest periods. Each image was shown for 500ms and was followed by a 1500ms inter-stimulus interval. Full-brain fMRI data were recorded with a volume repetition time of 2.5s, thus, a stimulus block was covered by roughly 9 volumes. This dataset has been repeatedly reanalyzed.
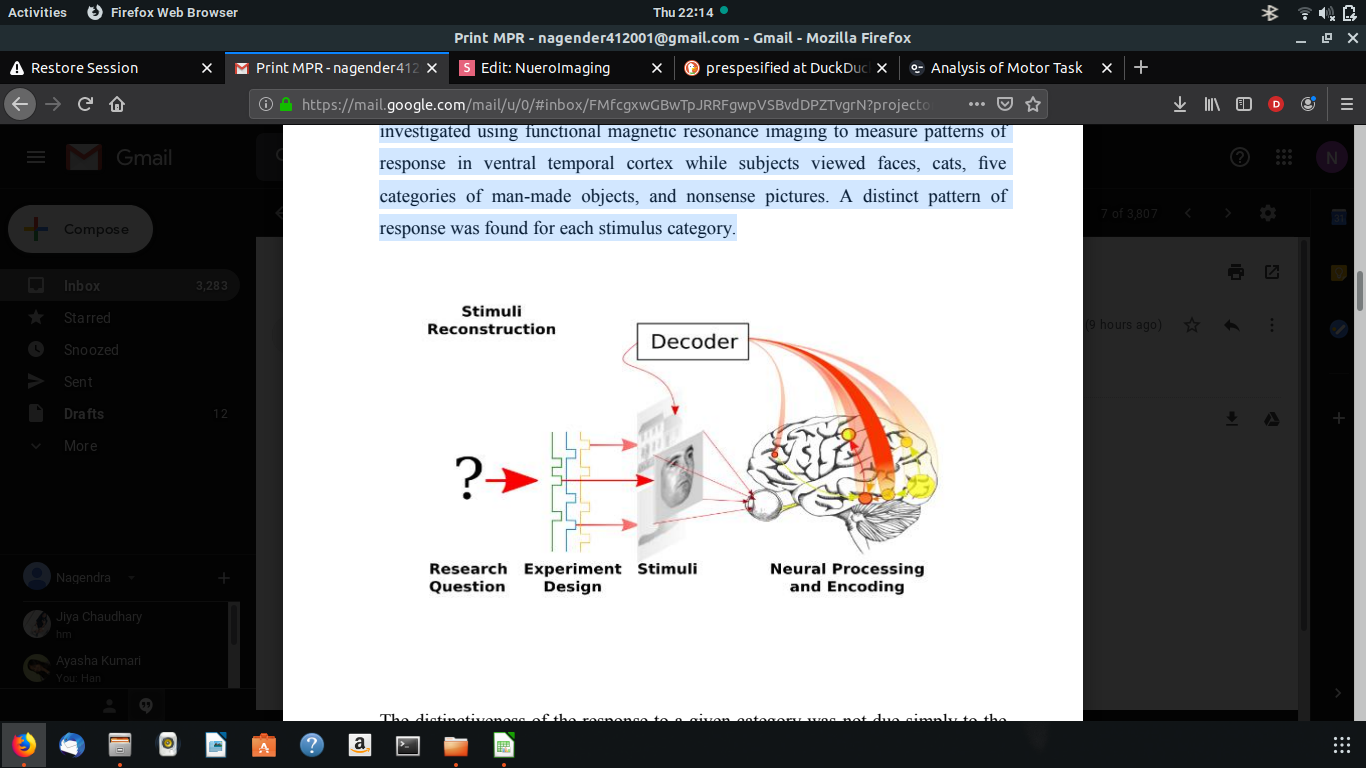
HAXBY EXPERIMENT
The functional architecture of the object vision pathway in the human brain was investigated using functional magnetic resonance imaging to measure patterns of response in ventral temporal cortex while subjects viewed faces, cats, five categories of man-made objects, and nonsense pictures. A distinct pattern of response was found for each stimulus category.
THANK-YOU
NueroImaging
By pranavkhurana
NueroImaging
Machine learning for neuroimaging with scikit-learn (Hackveda Project)
- 368



