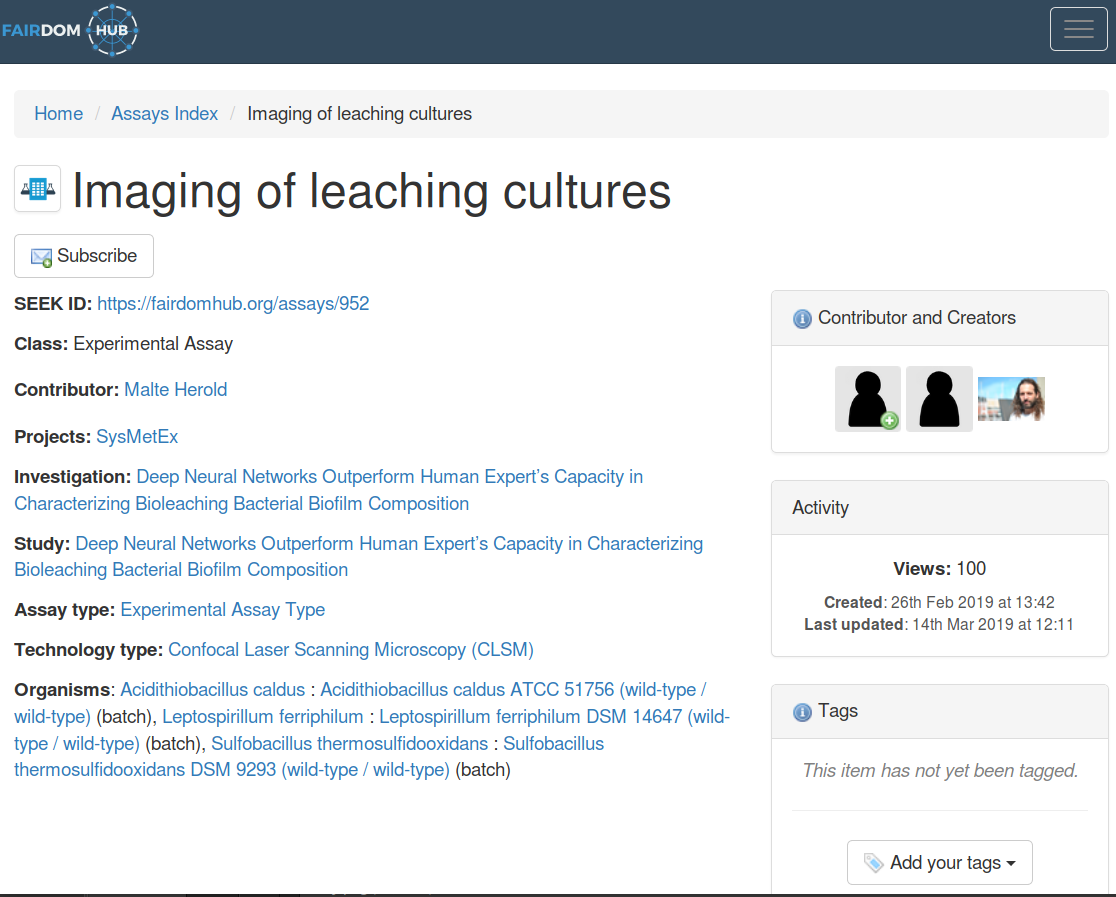
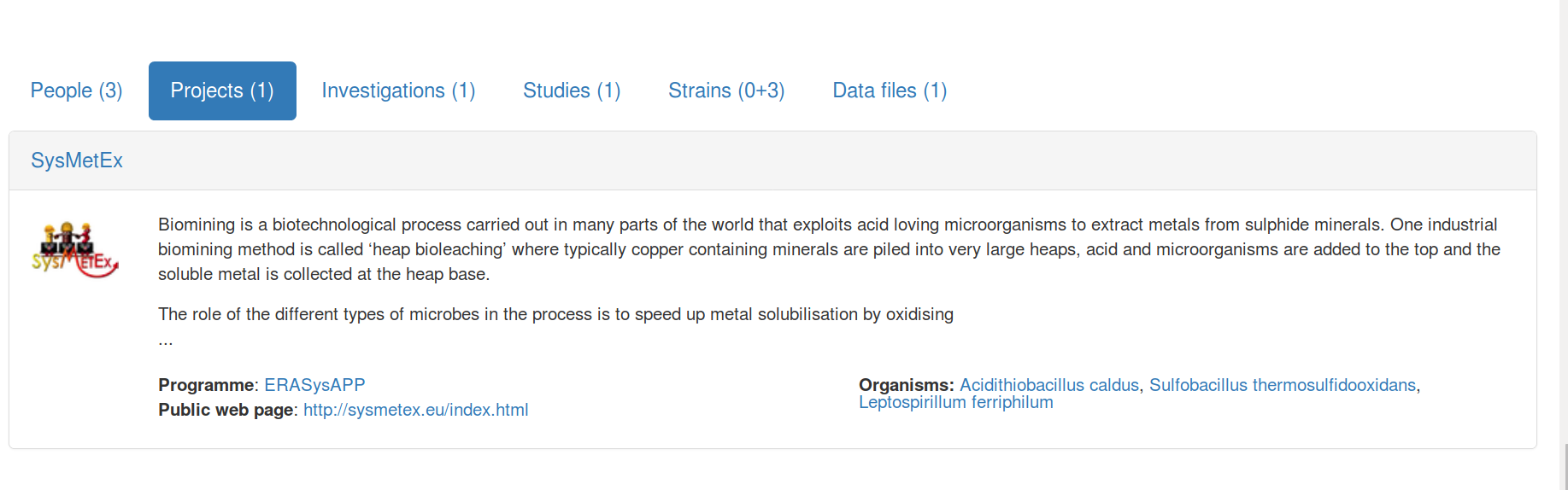
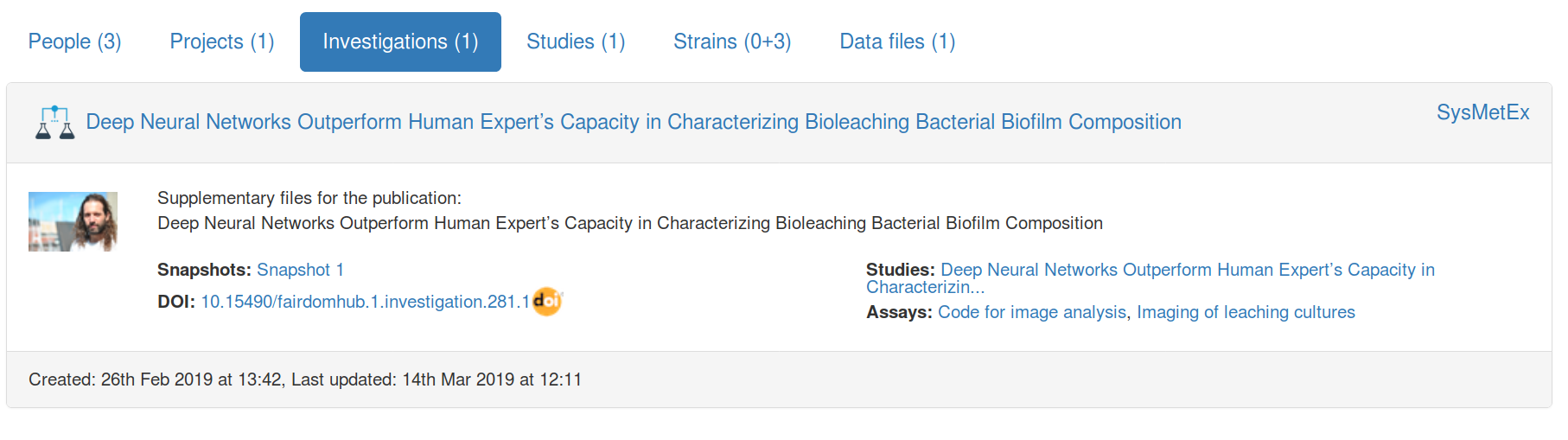
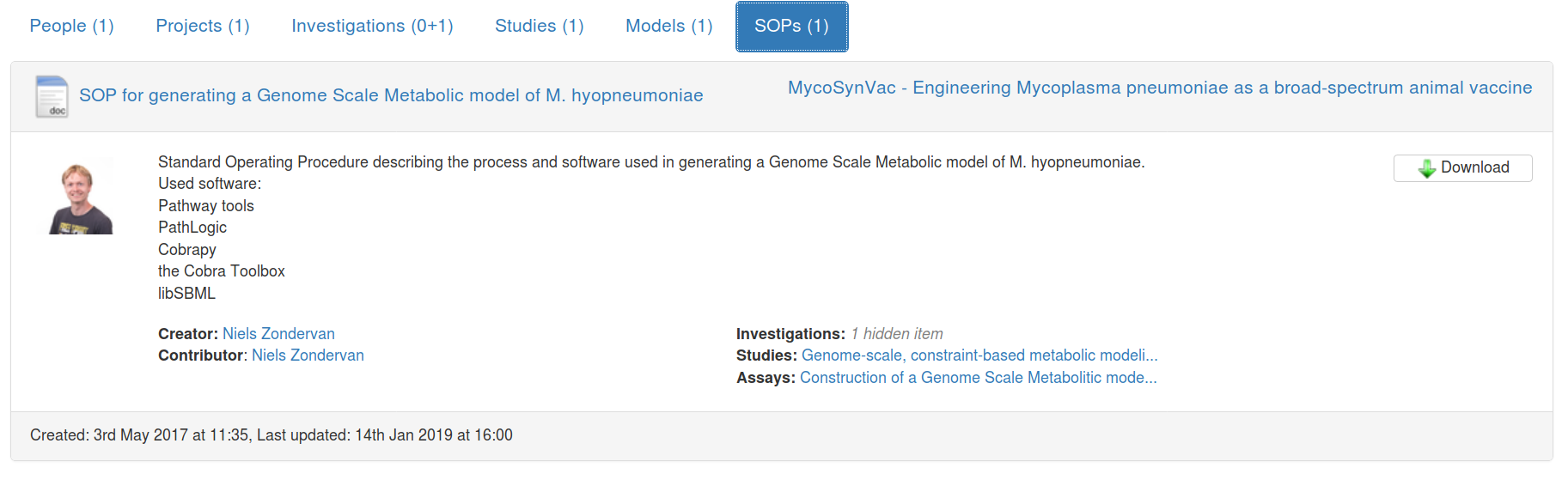
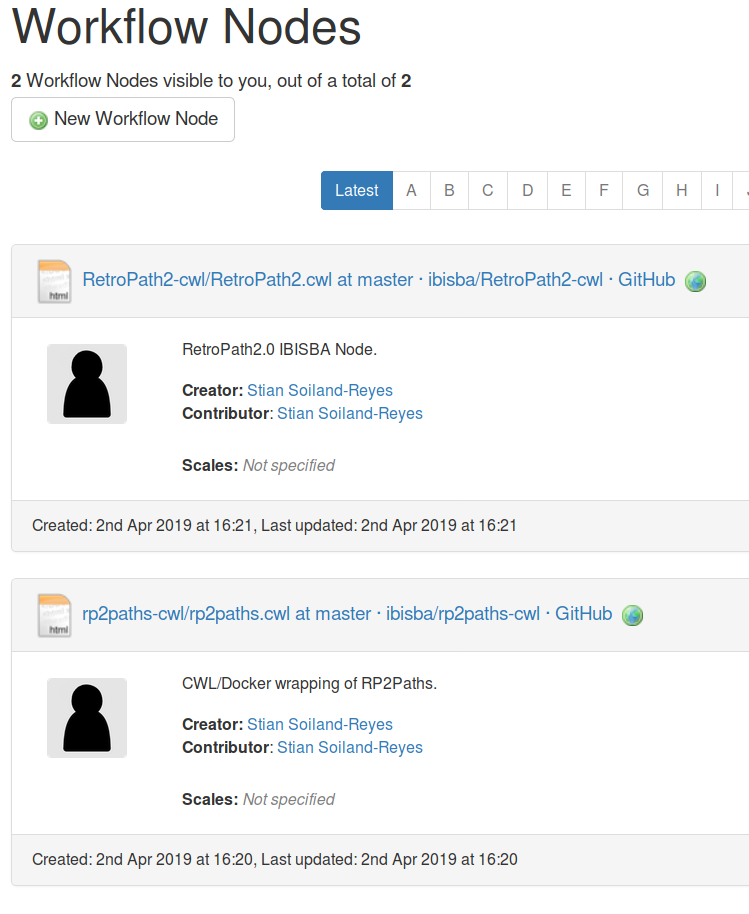
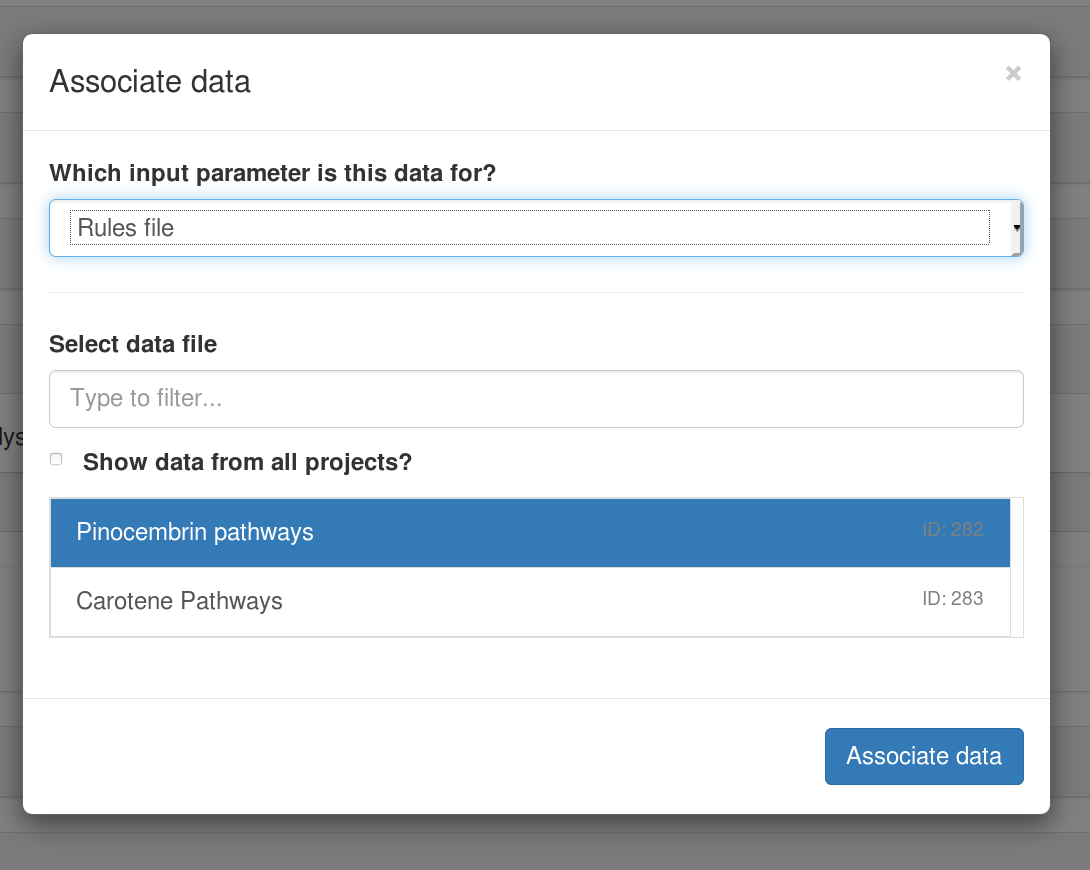
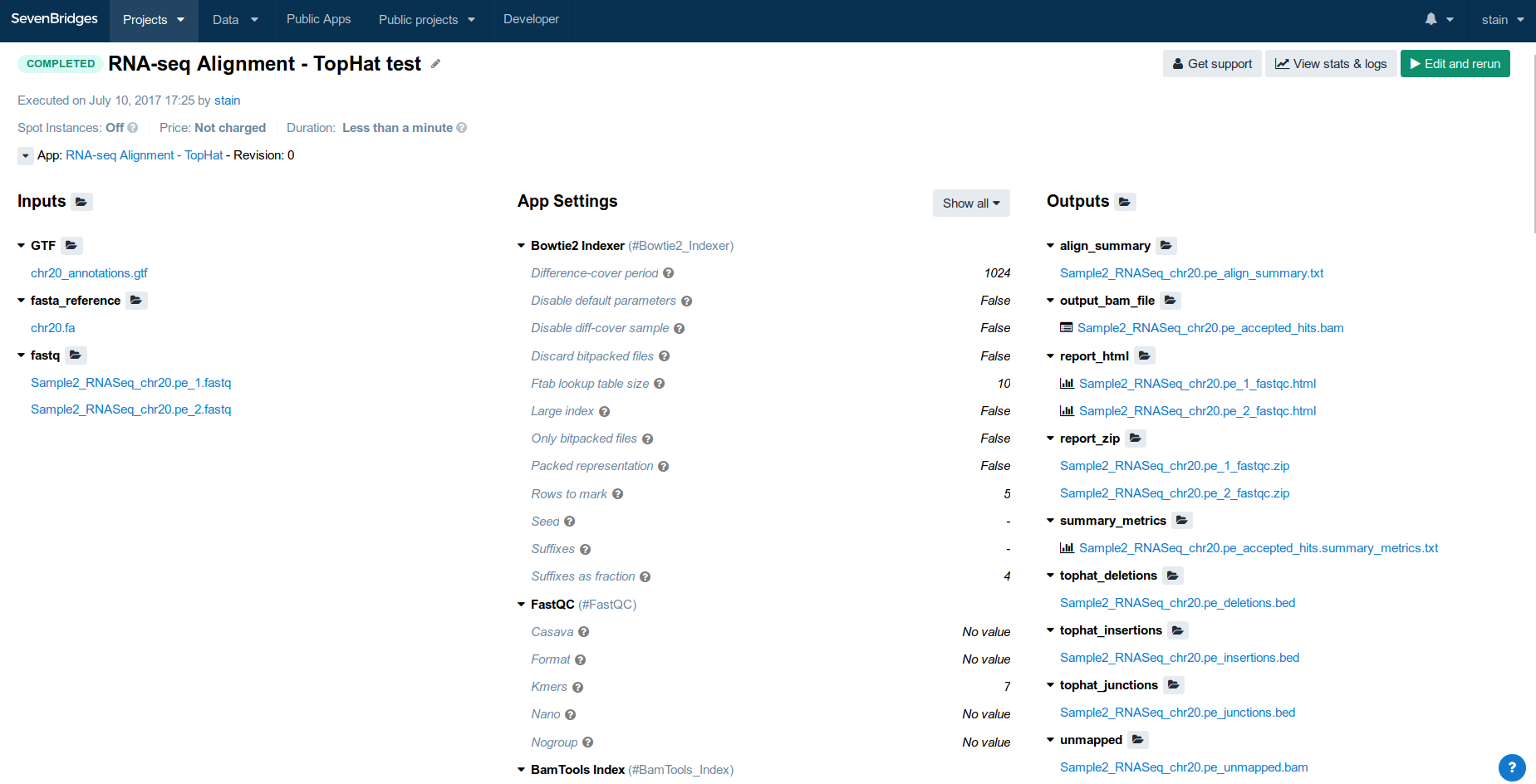
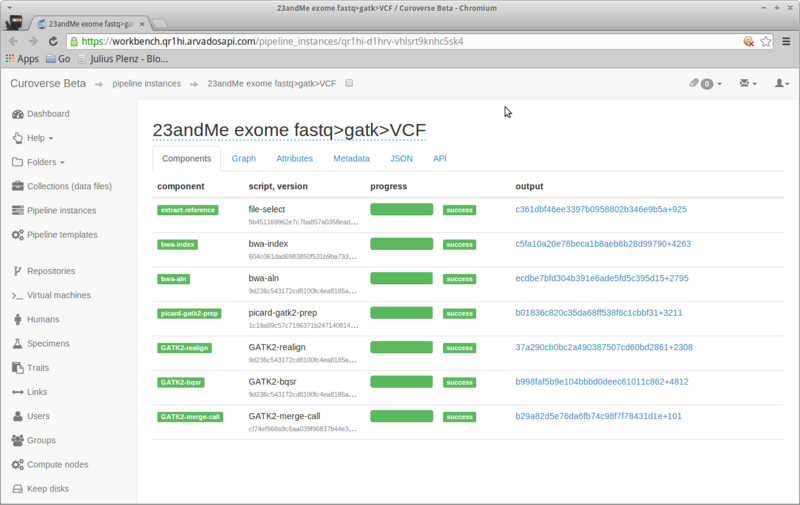
$ cwlprov --help
usage: cwlprov [-h] [--version] [--directory DIRECTORY] [--relative]
[--absolute] [--output OUTPUT] [--verbose] [--quiet] [--hints]
[--no-hints]
{validate,info,who,prov,inputs,outputs,run,runs,rerun,derived,runtimes}
...
cwlprov explores Research Objects containing provenance of Common Workflow
Language executions. <https://w3id.org/cwl/prov/>
commands:
{validate,info,who,prov,inputs,outputs,run,runs,rerun,derived,runtimes}
validate Validate the CWLProv Research Object
info show research object Metadata
who show Who ran the workflow
prov export workflow execution Provenance in PROV format
inputs list workflow/step Input files/values
outputs list workflow/step Output files/values
run show workflow Execution log
runs List all workflow executions in RO
rerun Rerun a workflow or step
derived list what was Derived from a data item, based on
activity usage/generation
runtimes calculate average step execution Runtimes