Benchmarking and methods development for single cell data
PhD defense
Almut Lütge
Zürich, 28.06.23


single cell (transcriptome) data



Cells are the basic unit of life
Cells are the basic unit of life

Development



Stem cell
Differentiated cells
Tissue


Cells are the basic unit of life

Development




Stem cell
Differentiated cells
Tissue



Disease
Cancer
Immune system



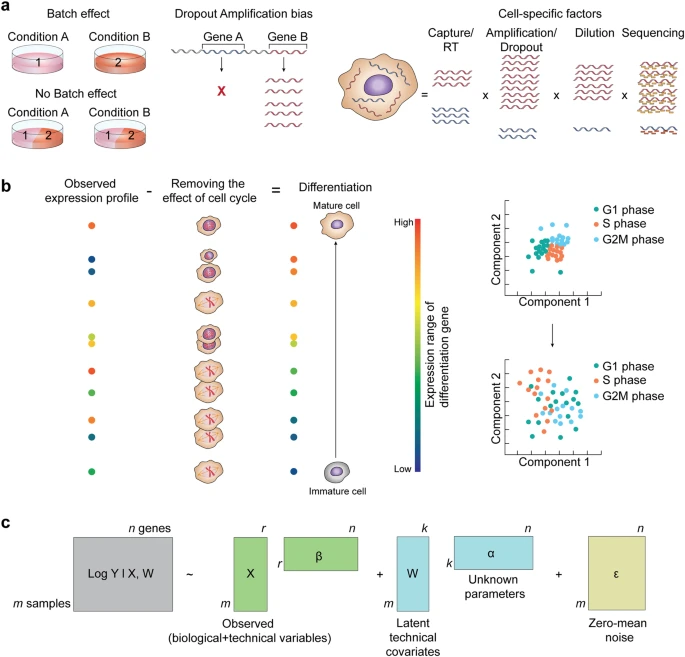
Cells are regulated at different molecular levels






Cells are regulated at different molecular levels






Cells are regulated at different molecular levels



Transcription





Cells are regulated at different molecular levels





Transcription
Translation

From reads to gene expression profiles
aatgctgcgctaatcgcgcgtatcgggatcatgccctagtggccccatattggcgtcaggtcgaacggatcttcggtgactccatgcattttcaggctcactgtggca
From reads to gene expression profiles
aatgctgcgctaatcgcgcgtatcgggatcatgccctagtggccccatattggcgtcaggtcgaacggatcttcggtgactccatgcattttcaggctcactgtggca

alignment, filtering, counting
From reads to gene expression profiles
aatgctgcgctaatcgcgcgtatcgggatcatgccctagtggccccatattggcgtcaggtcgaacggatcttcggtgactccatgcattttcaggctcactgtggca

alignment, filtering, counting

filtering,
QC, normalization

embedding
From reads to gene expression profiles
aatgctgcgctaatcgcgcgtatcgggatcatgccctagtggccccatattggcgtcaggtcgaacggatcttcggtgactccatgcattttcaggctcactgtggca

alignment, filtering, counting

filtering,
QC, normalization

embedding




clustering
trajectory
marker genes
differentiation
From reads to gene expression profiles
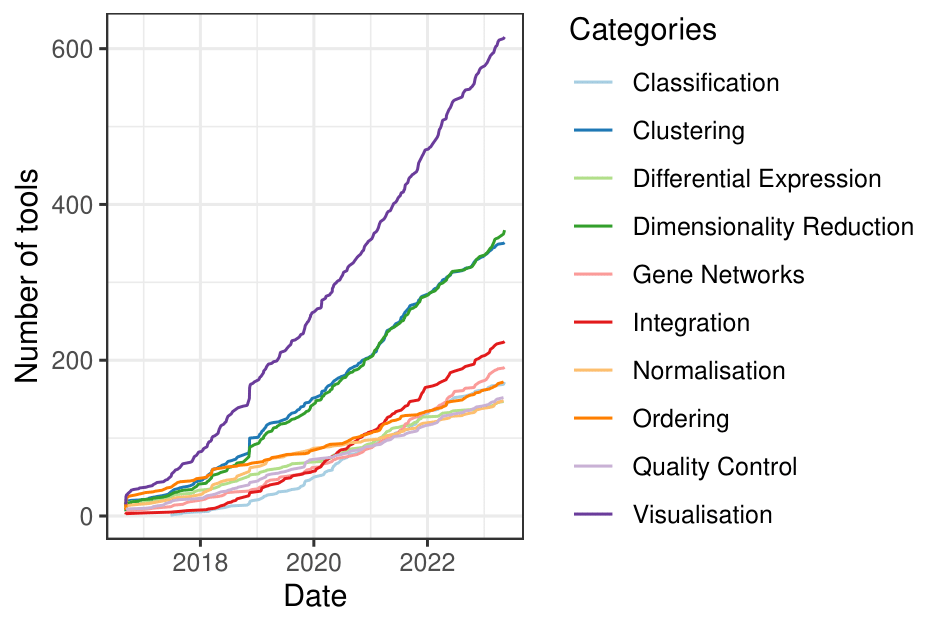
single cell tools are on the rise
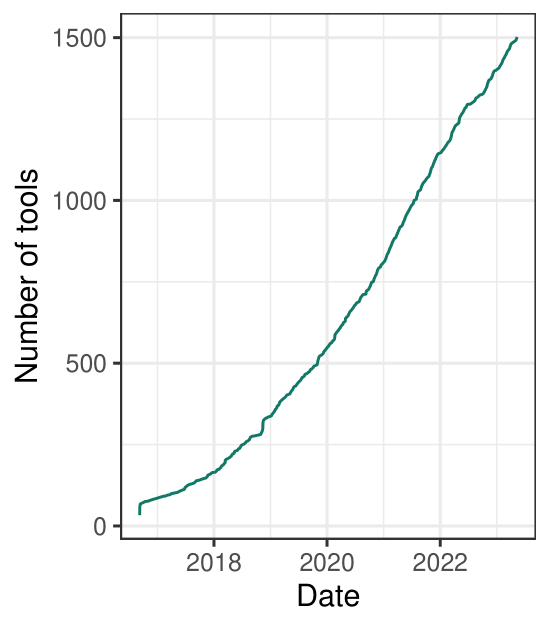
single cell tools are on the rise
Batch effects
Batch effects: Definition
Differences between data sets [..][that] occur due to uncontrolled variability in experimental factors (Lun, 2019)
Batch effects: Definition
Differences between data sets [..][that] occur due to uncontrolled variability in experimental factors (Lun, 2019)
- context-dependent
Batch effects: Definition
Differences between data sets [..][that] occur due to uncontrolled variability in experimental factors (Lun, 2019)
- context-dependent
- risk to mask biological signals or create spurious pattern
Example: No batch effect












Batch 1
Batch 2
Example: No batch effect












Batch 1
Batch 2
tsne1
tsne2
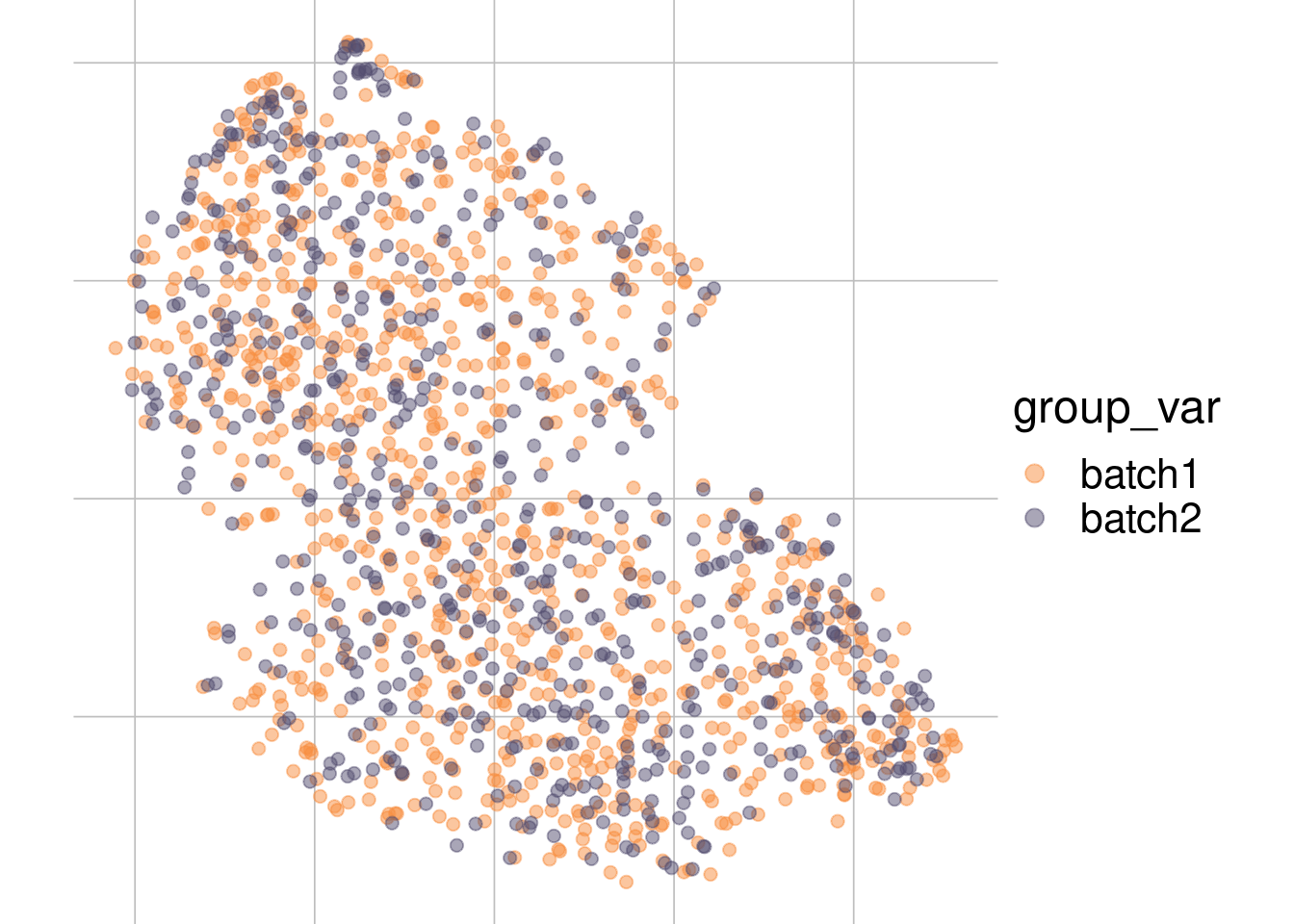
Example: batch effect












Batch 1
Batch 2
Example: batch effect












Batch 1
Batch 2
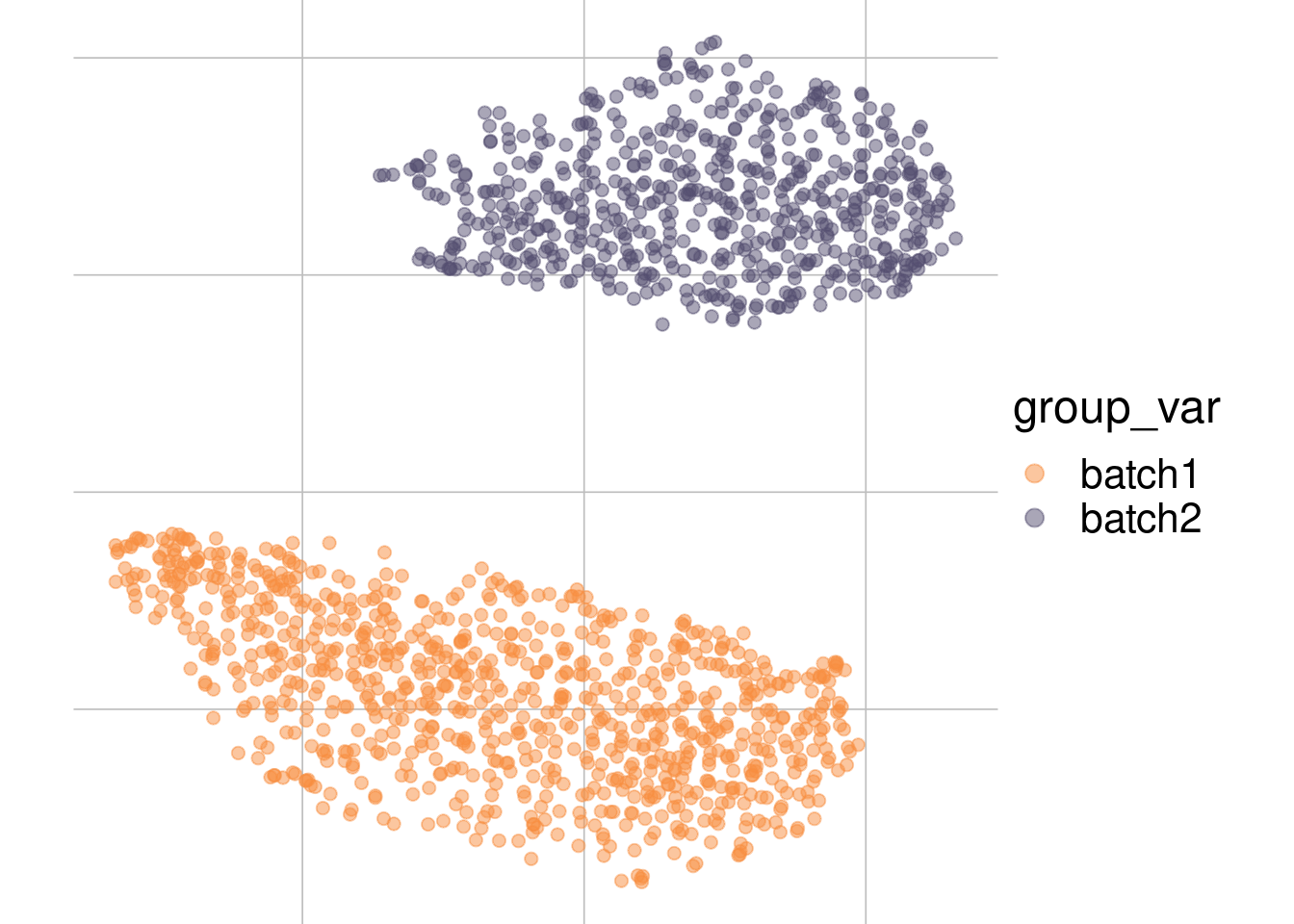
tsne1
tsne2
Example: cell types + No batch effect






Batch 1











Batch 2




Example: cell types + No batch effect






Batch 1











Batch 2




tsne1
tsne2






Batch 1











Batch 2




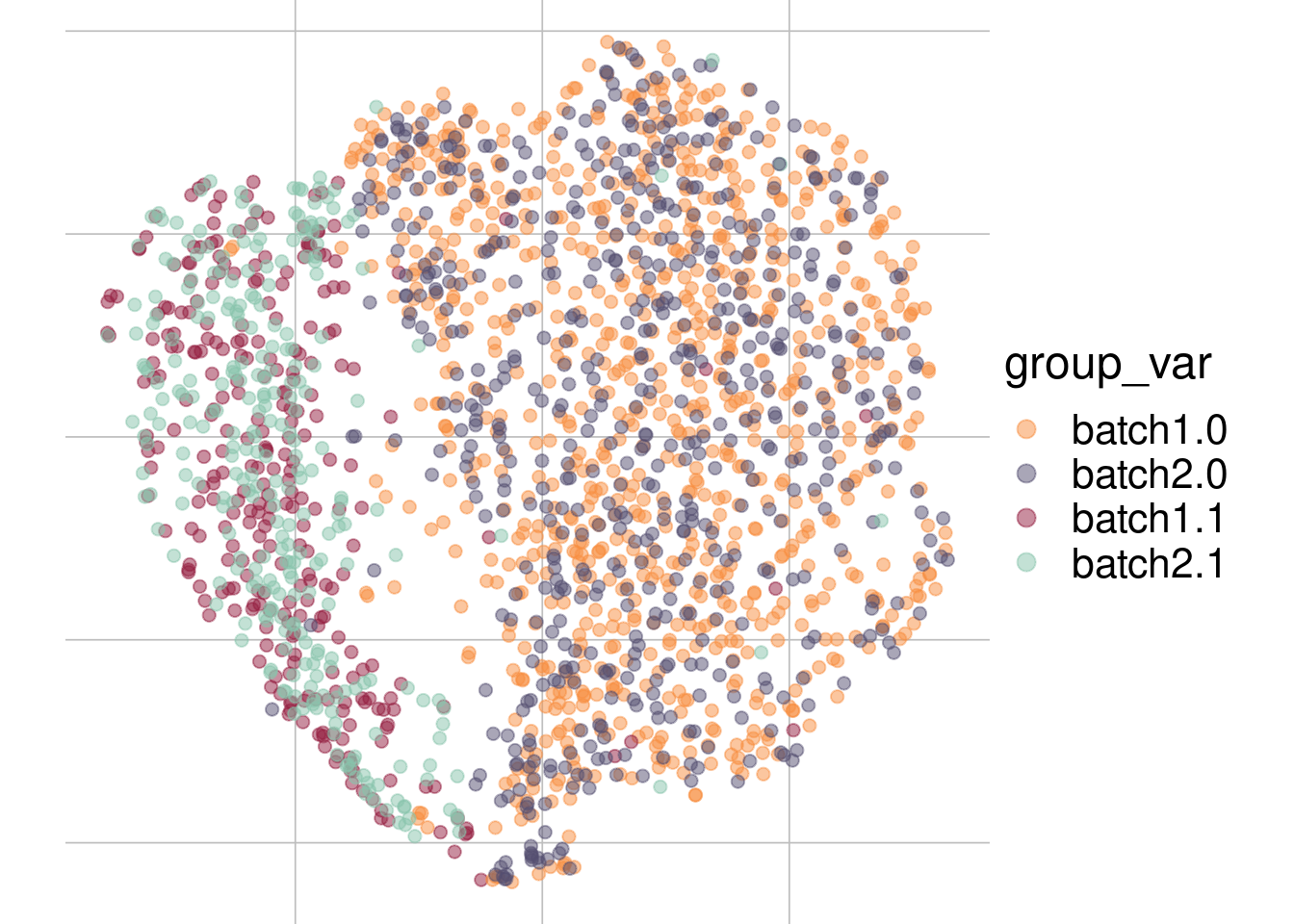
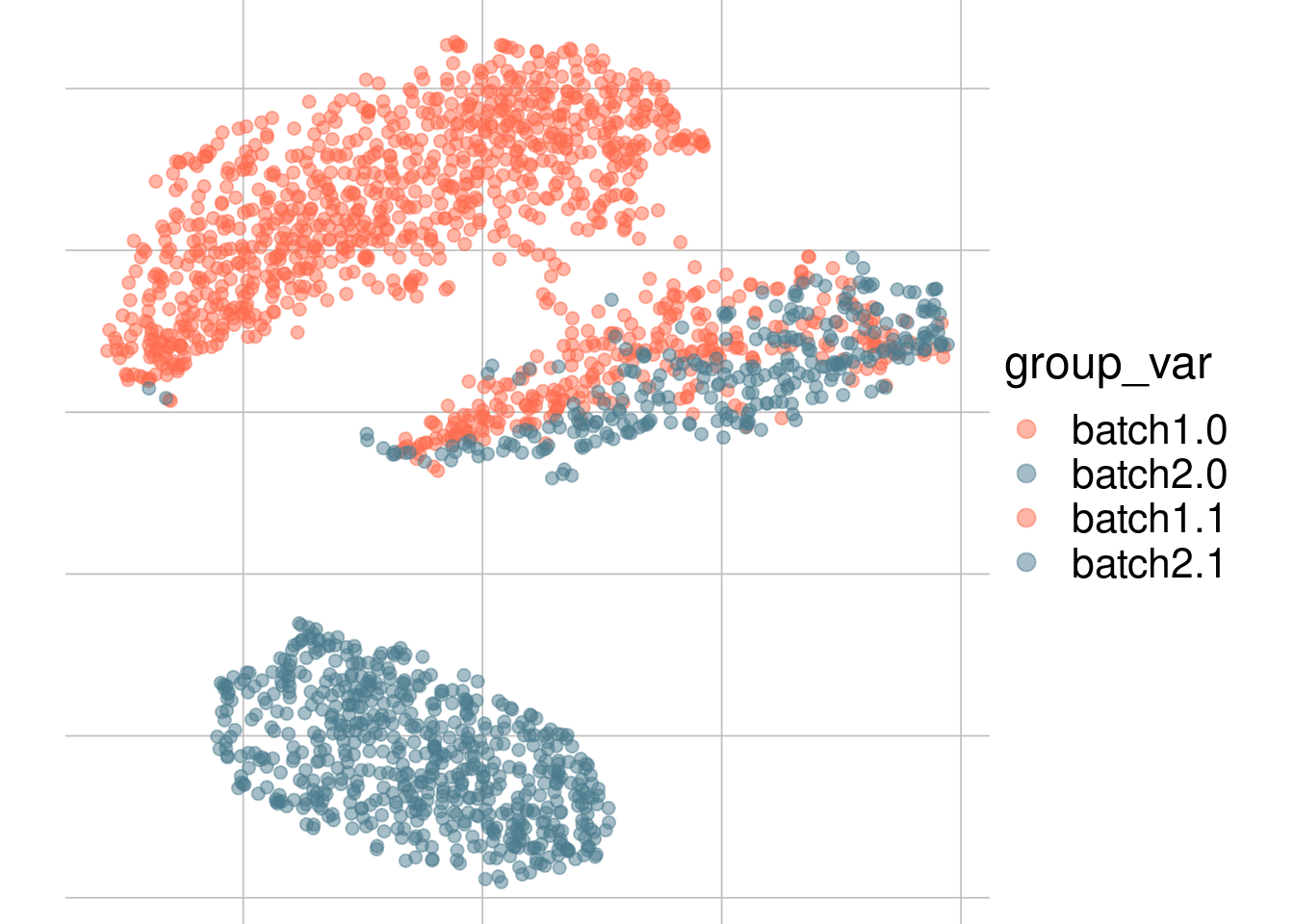
Example: cell types + batch effect






Batch 1











Batch 2




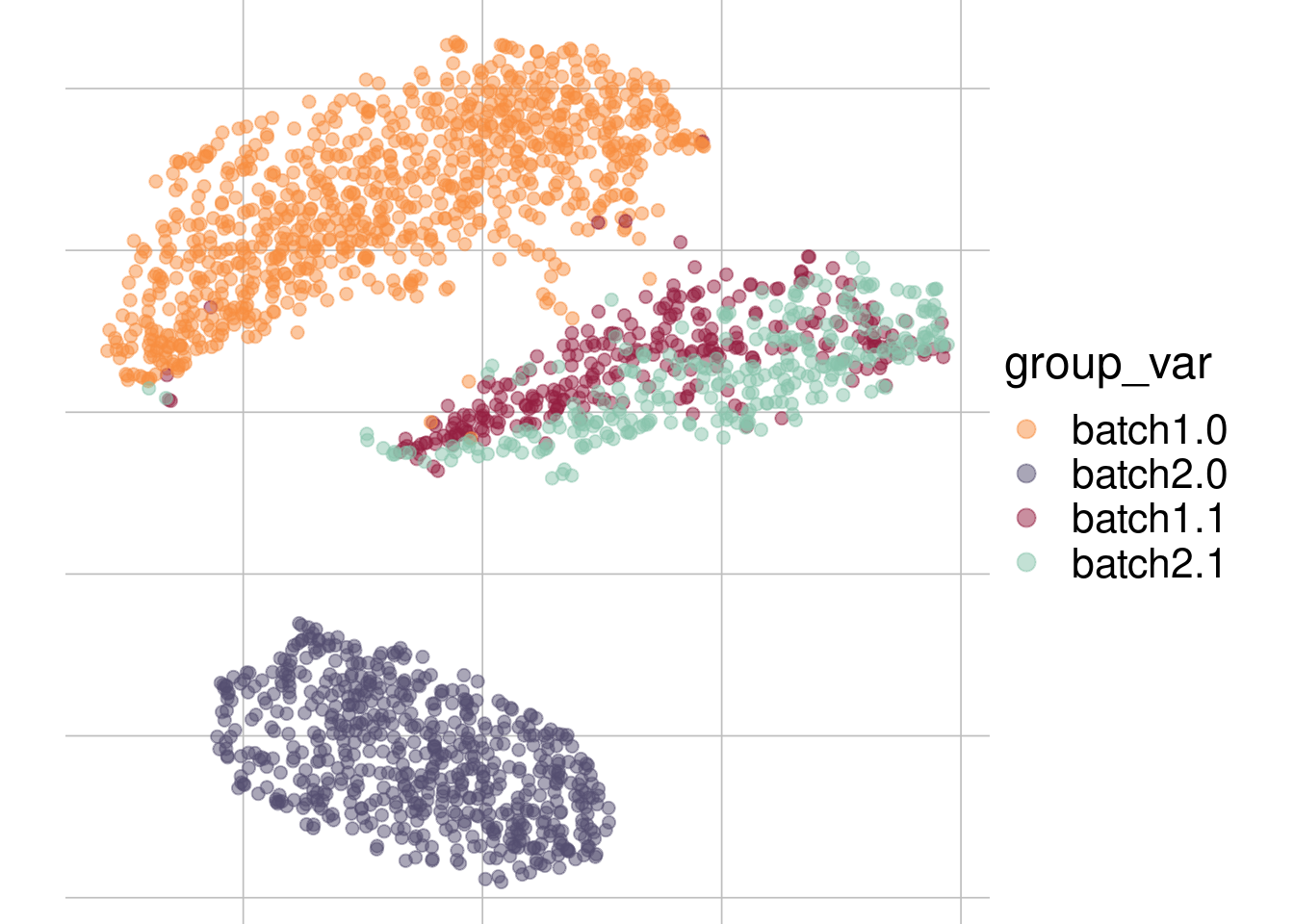
tsne1
tsne2
Example: cell types + batch effect
Example: batch effect






Batch 1











Batch 2




tsne1
tsne2
tsne1
tsne2
batch Integration (correction)






Batch 2










Batch 1





Common embedding





















dim1
dim2
single cell tools are on the rise

Batch mixing metrics:
How to quantify batch effects? Comparison of different batch mixing metrics
How to quantify batch effects?
- quantify "mixing" of batches
- Different levels: global, cell type, cell
How to quantify batch effects?
- quantify "mixing" of batches
- Different levels: global, cell type, cell
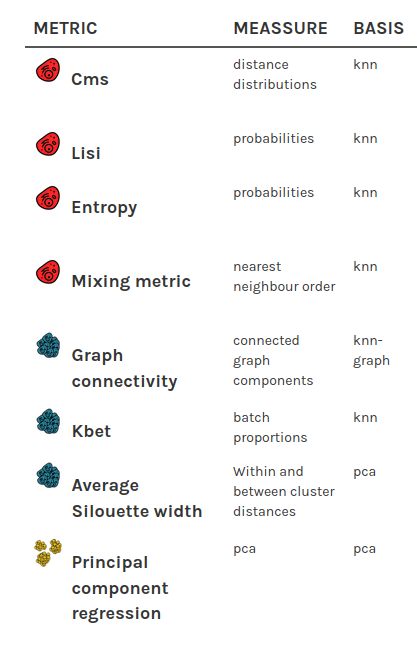
How to quantify batch effects?
- quantify "mixing" of batches
- Different levels: global, cell type, cell


Cellspecific mixing score (cms): No batch effect
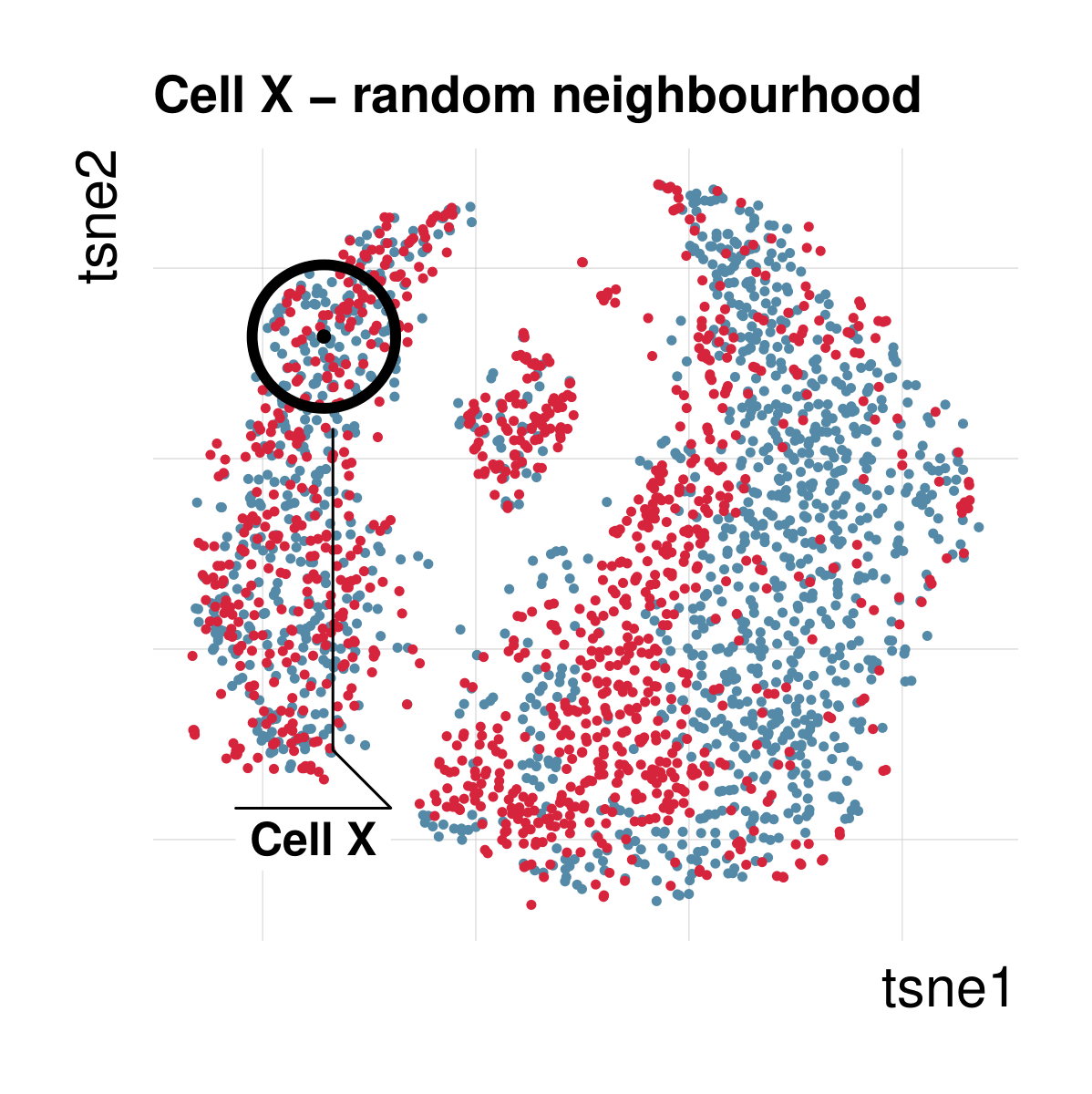
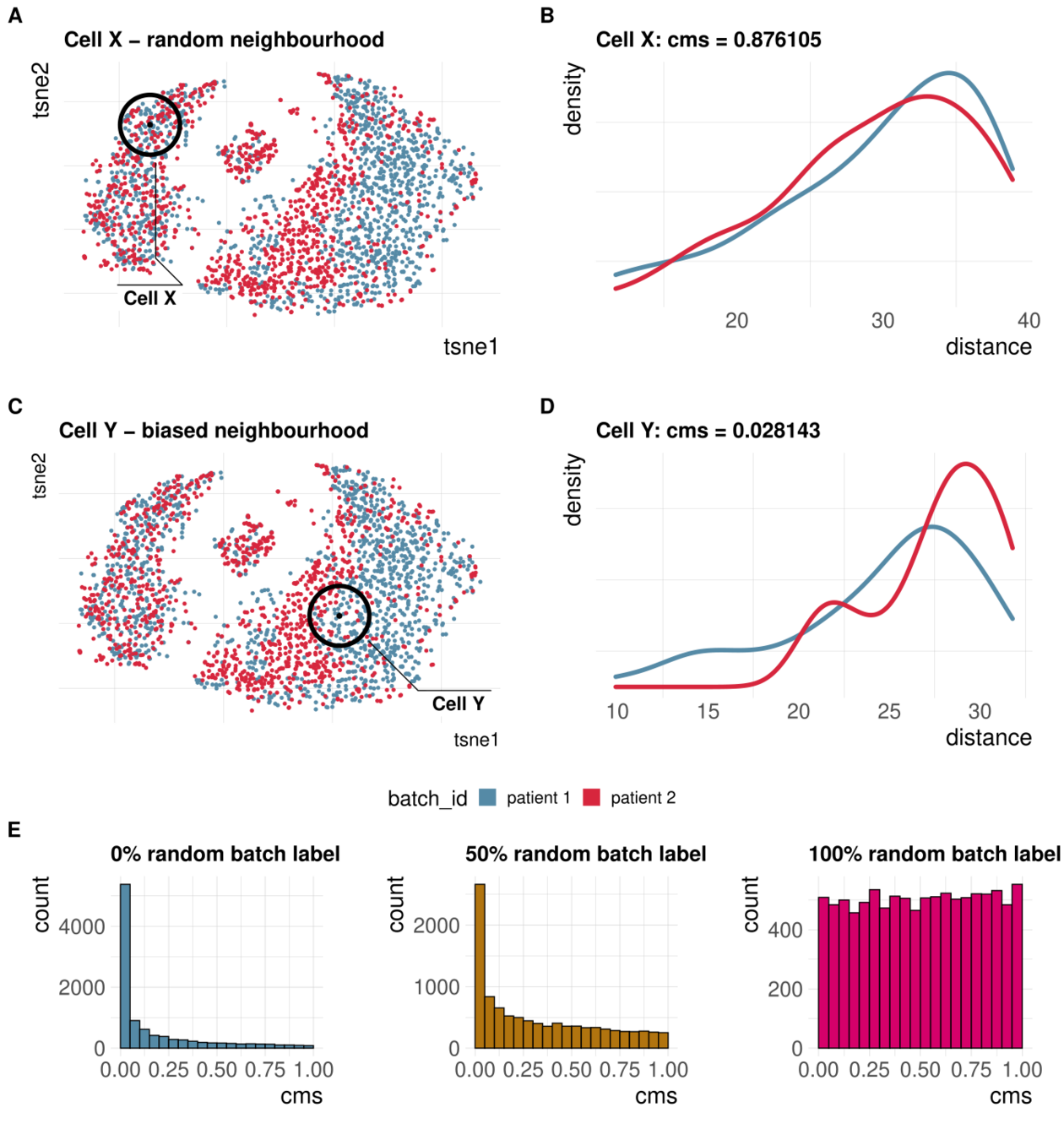
Cellspecific mixing score (cms): No batch effect

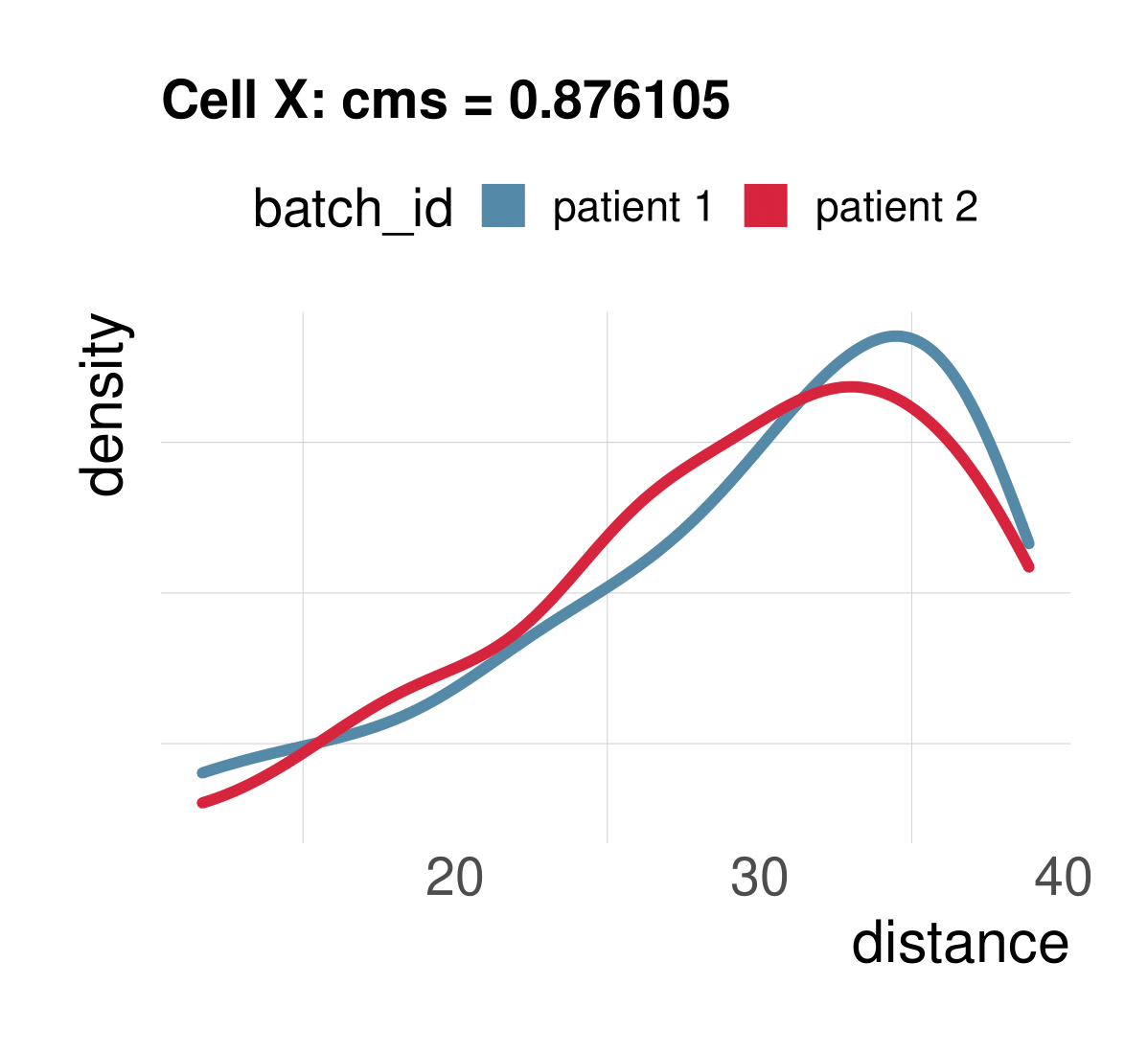

Cellspecific mixing score (cms): No batch effect
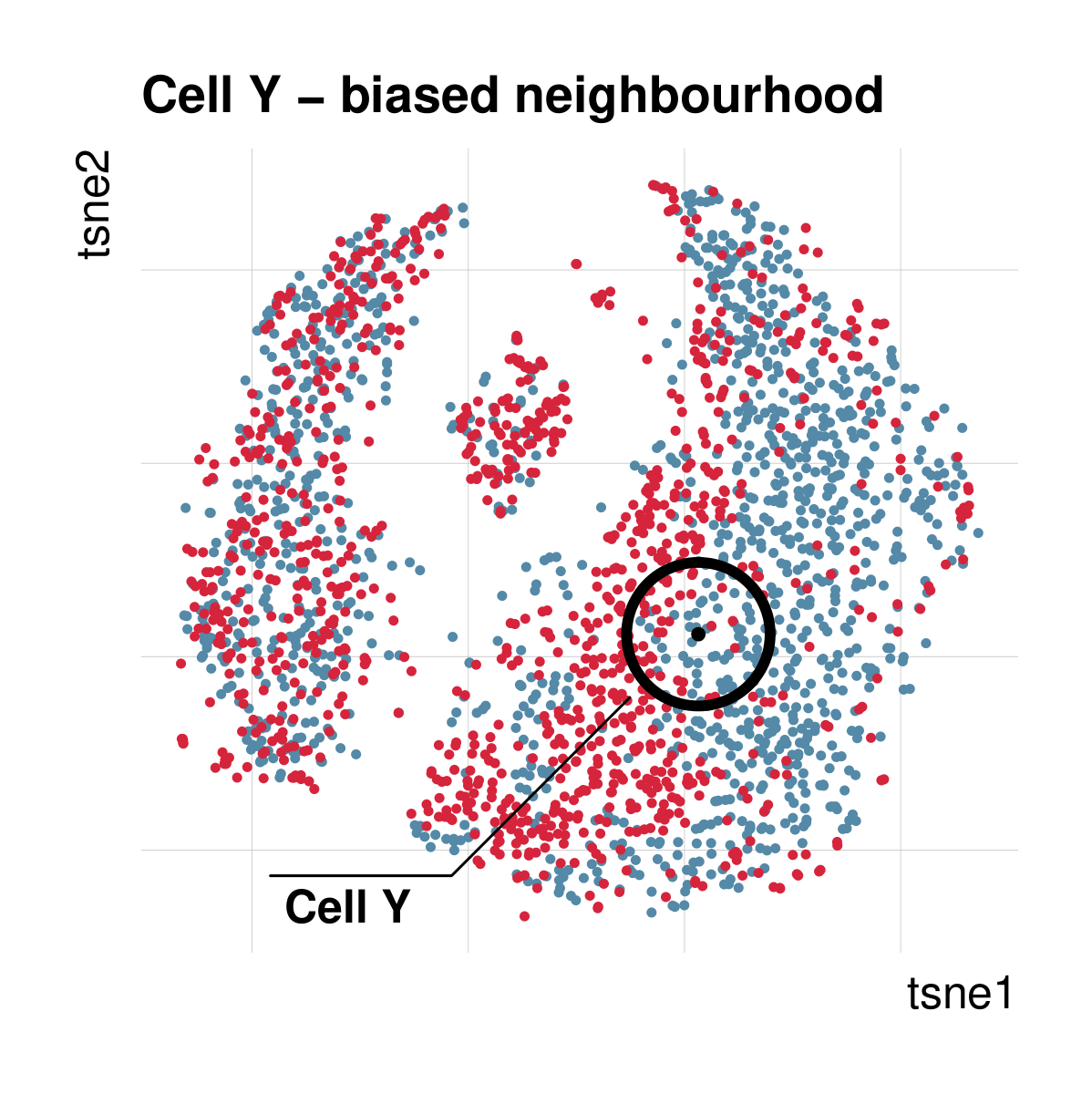

Cellspecific mixing score (cms): No batch effect

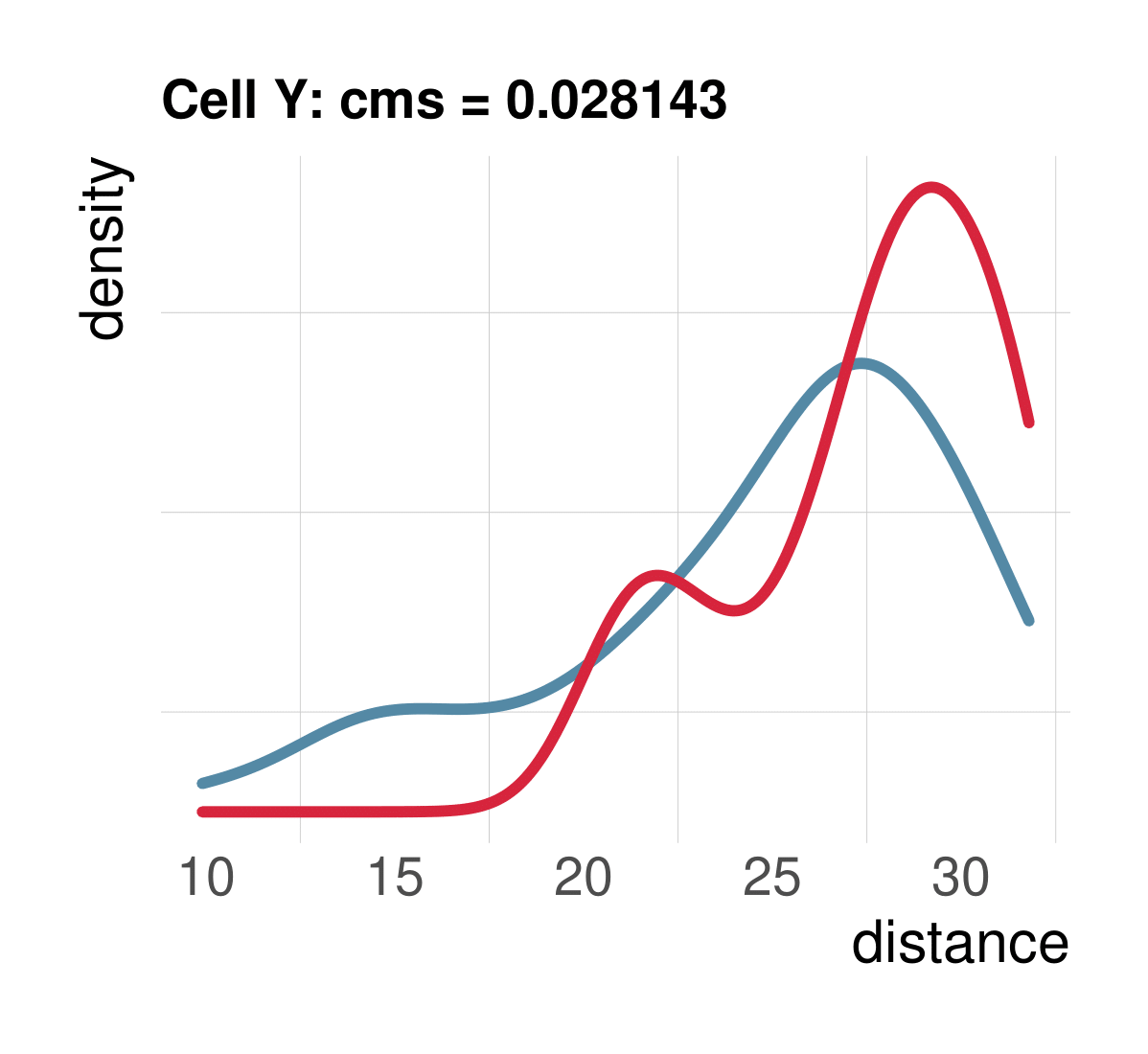

CMS scales with batch randomness

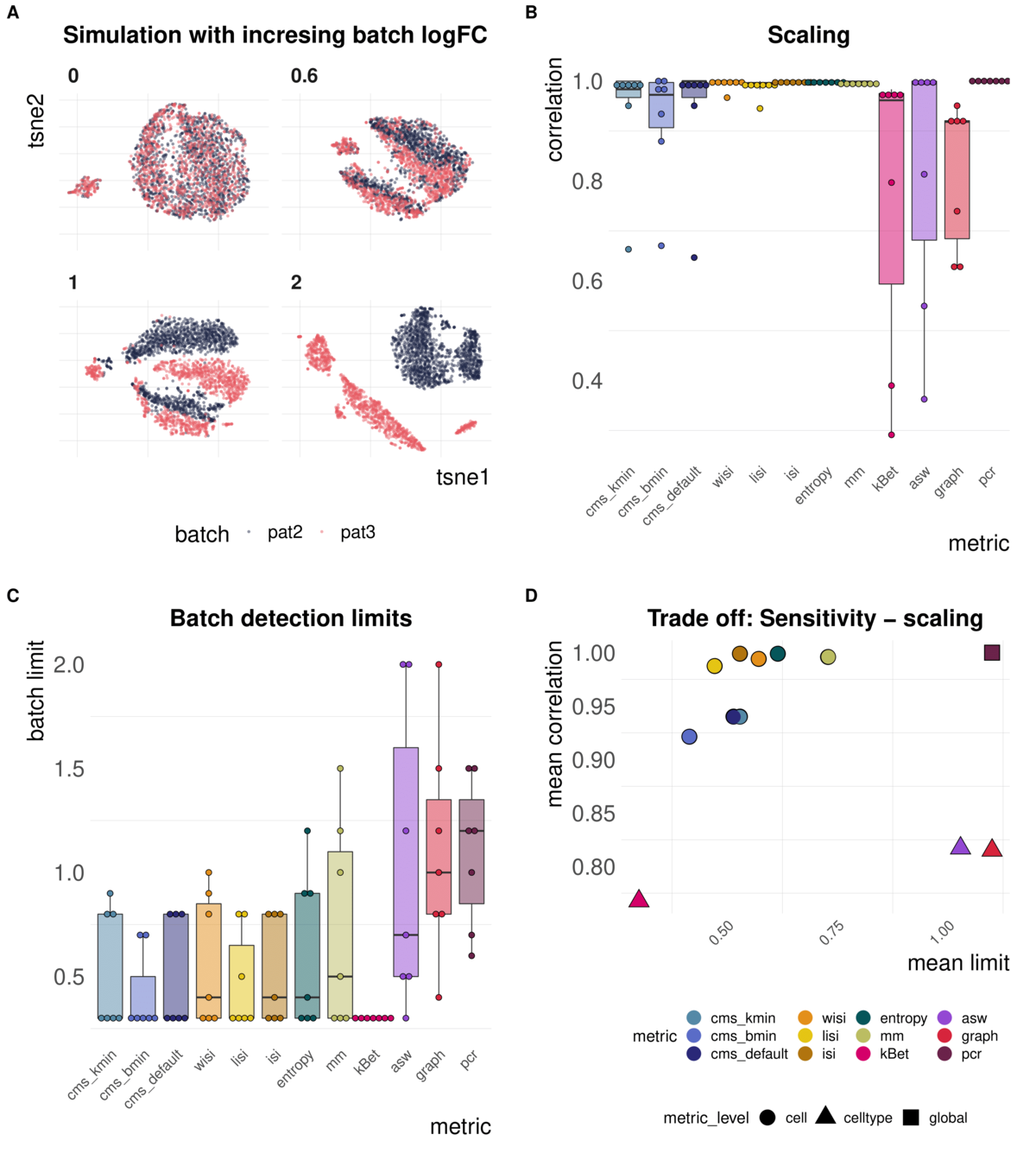
Task 1: Scaling and sensitivity
Aim: Test whether metrics scale with (synthetic) batch strength; Estimate lower limit of batch detection
Spearman correlation of metrics with the batch logFC in simulation series on the same dataset; Minimal batch logFC that is recognized from the metrics as batch effect
Task 1: increasing batch strength
Task 1: increasing batch strength
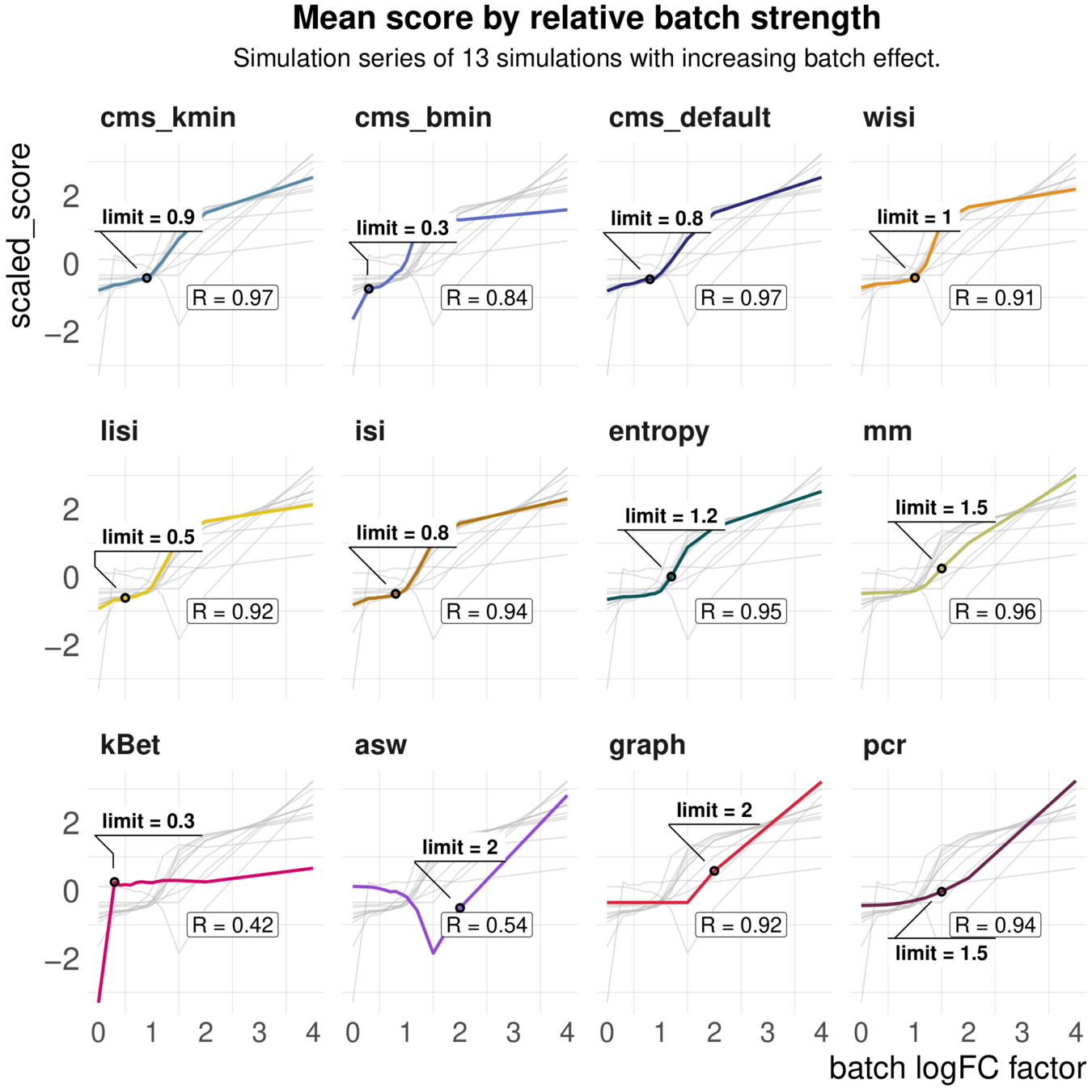


Metrics vary in their batch detection ranges

batch mixing metrics are context-dependent
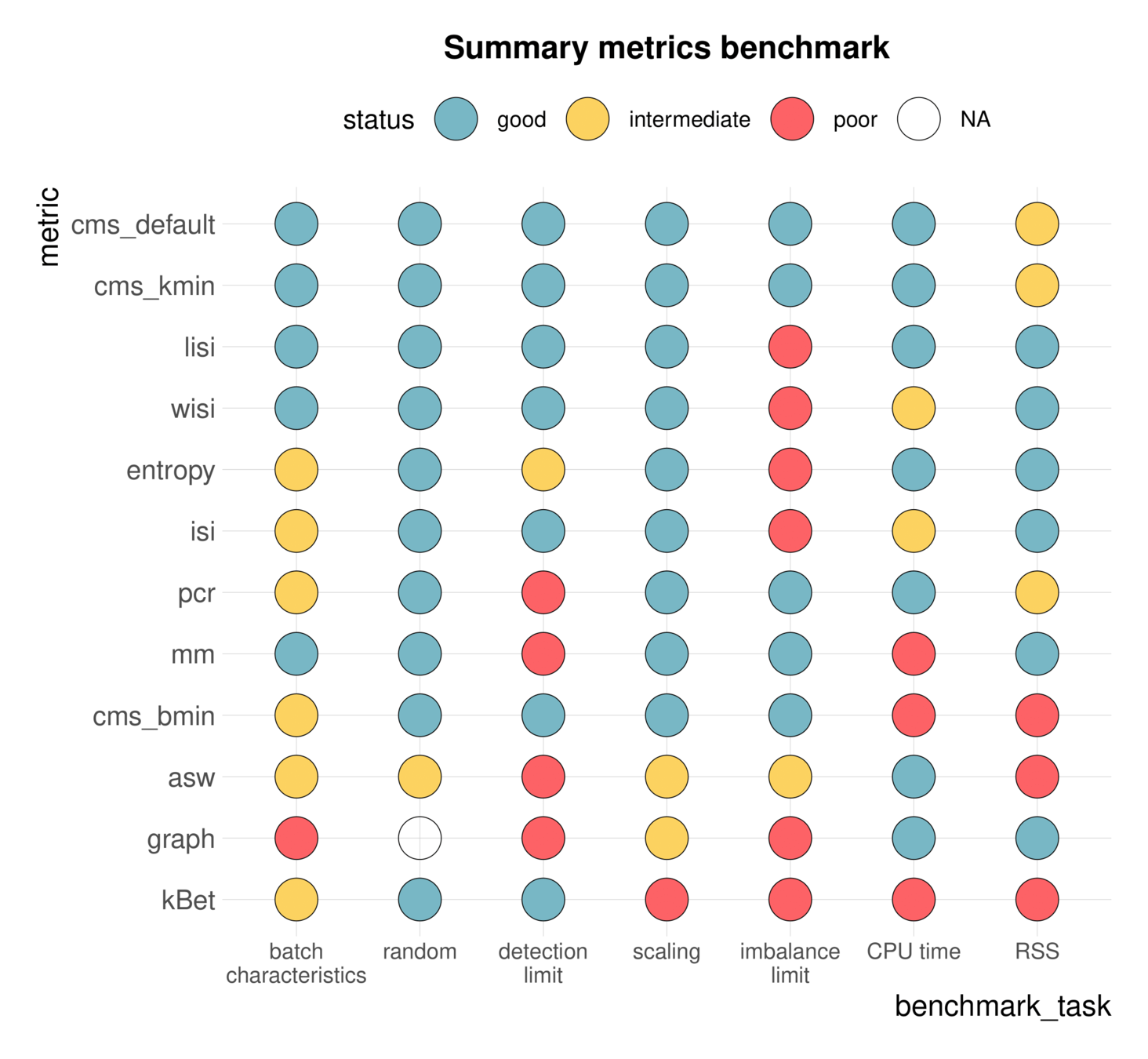
Benchmarking
Systematic performance comparisons
Datasets


Systematic performance comparisons
Datasets


Methods
Systematic performance comparisons
Datasets


Methods
Metrics

Systematic performance comparisons
Datasets


Methods
Metrics



benchmarks to guide method choice

The self assessment trap

The self assessment trap


1. cms_default 2. cms_kmin 3. lisi
The self assessment trap


1. cms_default 2. cms_kmin 3. lisi

Norel et al, 2011
Benchmarking results can be ambiguous
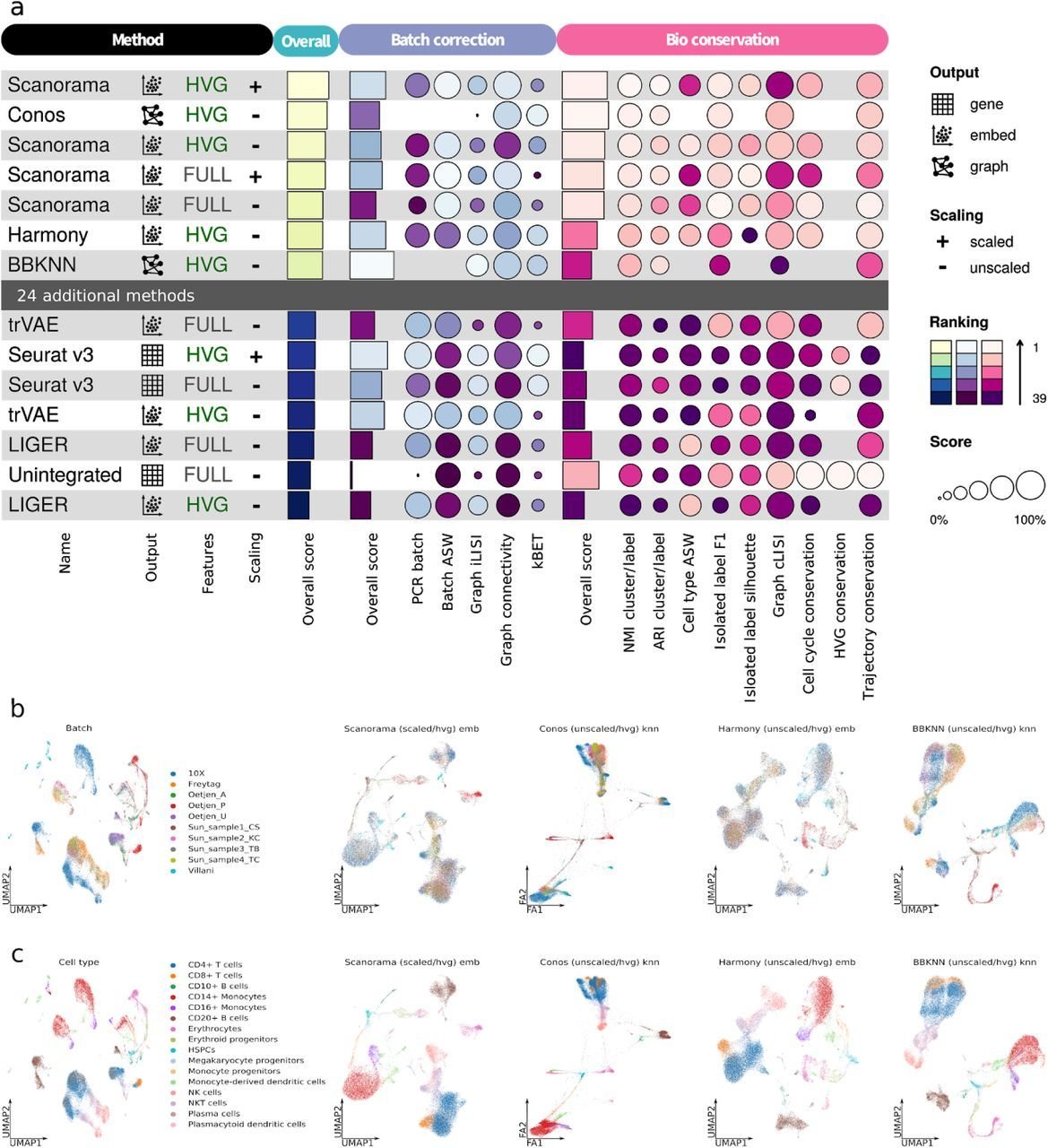
Luecken et al., 2021

Different benchmarks - different conclusions
- Scanorama
- Conos
- Harmony
- Limma
- Combat



- Liger
- Seurat
- Harmony
-
- Seurat
- Harmony
- Scanorama
- Liger
- TrVAE
- Seurat

"benchmarking [..] is comparable to asking how good a baseball player is by testing how quickly he or she hits or runs under very controlled circumstances." Kasper Lage, 2020
Do benchmarks reflect reality?
"benchmarking [..] is comparable to asking how good a baseball player is by testing how quickly he or she hits or runs under very controlled circumstances." Kasper Lage, 2020

Robert Lewandowski
max. speed: 32.71 km/h

Timo Werner
max. speed: 34.1 km/h
Do benchmarks reflect reality?
"benchmarking [..] is comparable to asking how good a baseball player is by testing how quickly he or she hits or runs under very controlled circumstances." Kasper Lage, 2020

Robert Lewandowski
max. speed: 32.71 km/h
passes: 765

Timo Werner
max. speed: 34.1 km/h
passes: 660
Do benchmarks reflect reality?
"benchmarking [..] is comparable to asking how good a baseball player is by testing how quickly he or she hits or runs under very controlled circumstances." Kasper Lage, 2020

Robert Lewandowski
max. speed: 32.71 km/h
passes: 765
goals: 23

Timo Werner
max. speed: 34.1 km/h
passes: 660
goals: 9
Do benchmarks reflect reality?
status quo:
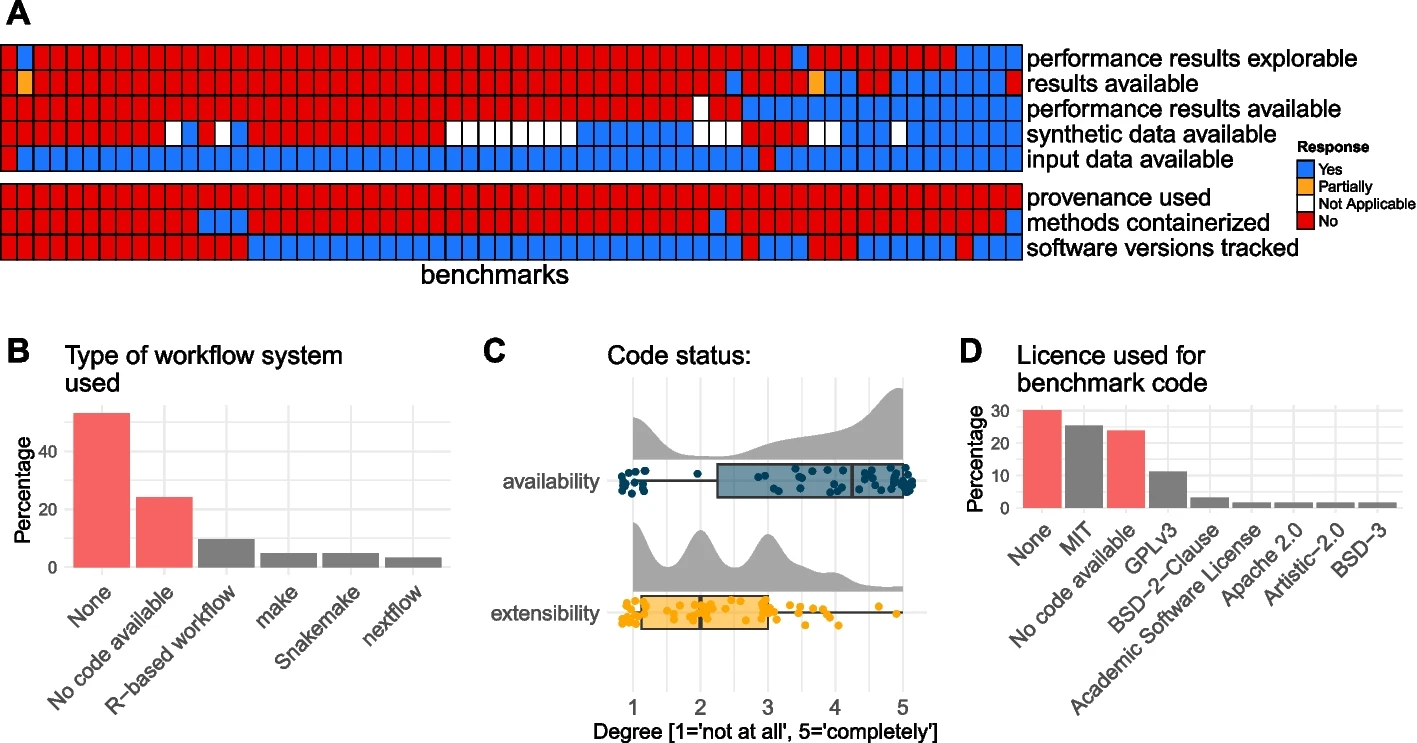
Meta-analysis of 62 method benchmarks in the field of single cell omics


62 single cell omics method benchmarks
62 single cell omics method benchmarks
Meta-analysis:
-
Title
-
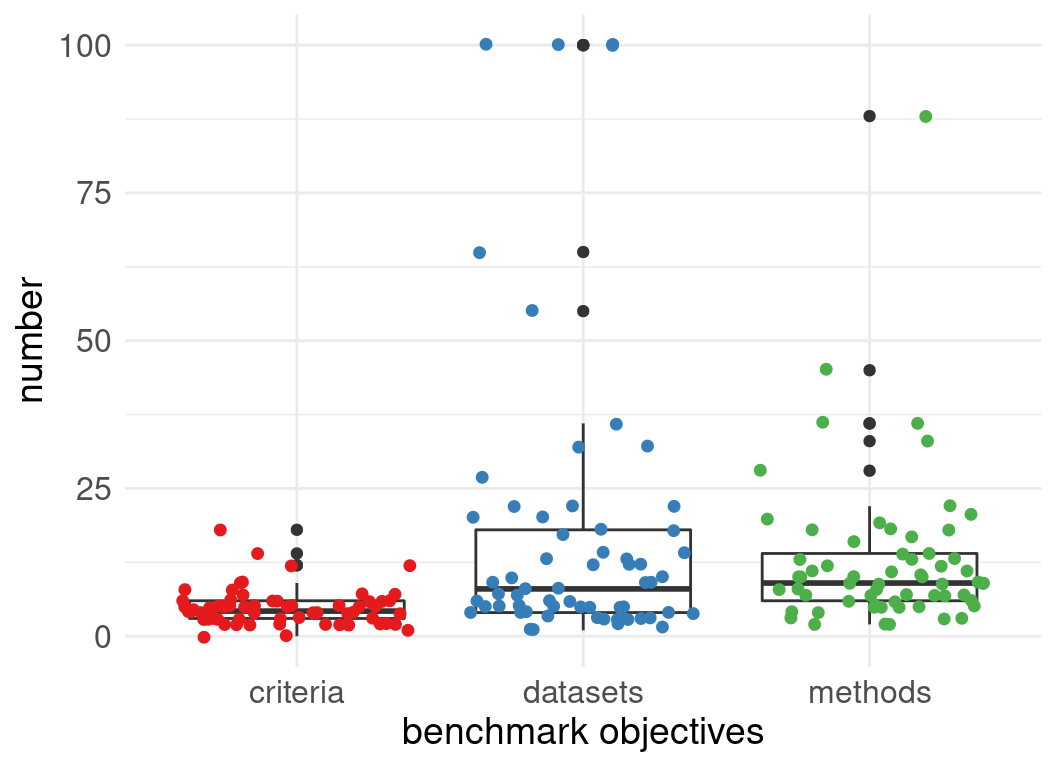
Number of datasets used in evaluations:
-
Number of methods evaluated:
-
Degree to which authors are neutral:
...
22. Type of workflow system used:
62 single cell omics method benchmarks
2 reviewer per benchmark
Meta-analysis:
-
Title
-
Number of datasets used in evaluations:
-
Number of methods evaluated:
-
Degree to which authors are neutral:
...
22. Type of workflow system used:
62 single cell omics method benchmarks
2 reviewer per benchmark
Meta-analysis:
-
Title
-
Number of datasets used in evaluations:
-
Number of methods evaluated:
-
Degree to which authors are neutral:
...
22. Type of workflow system used:
independent harmonization of responses
62 single cell omics method benchmarks
2 reviewer per benchmark
Meta-analysis:
-
Title
-
Number of datasets used in evaluations:
-
Number of methods evaluated:
-
Degree to which authors are neutral:
...
22. Type of workflow system used:
independent harmonization of responses

summaries
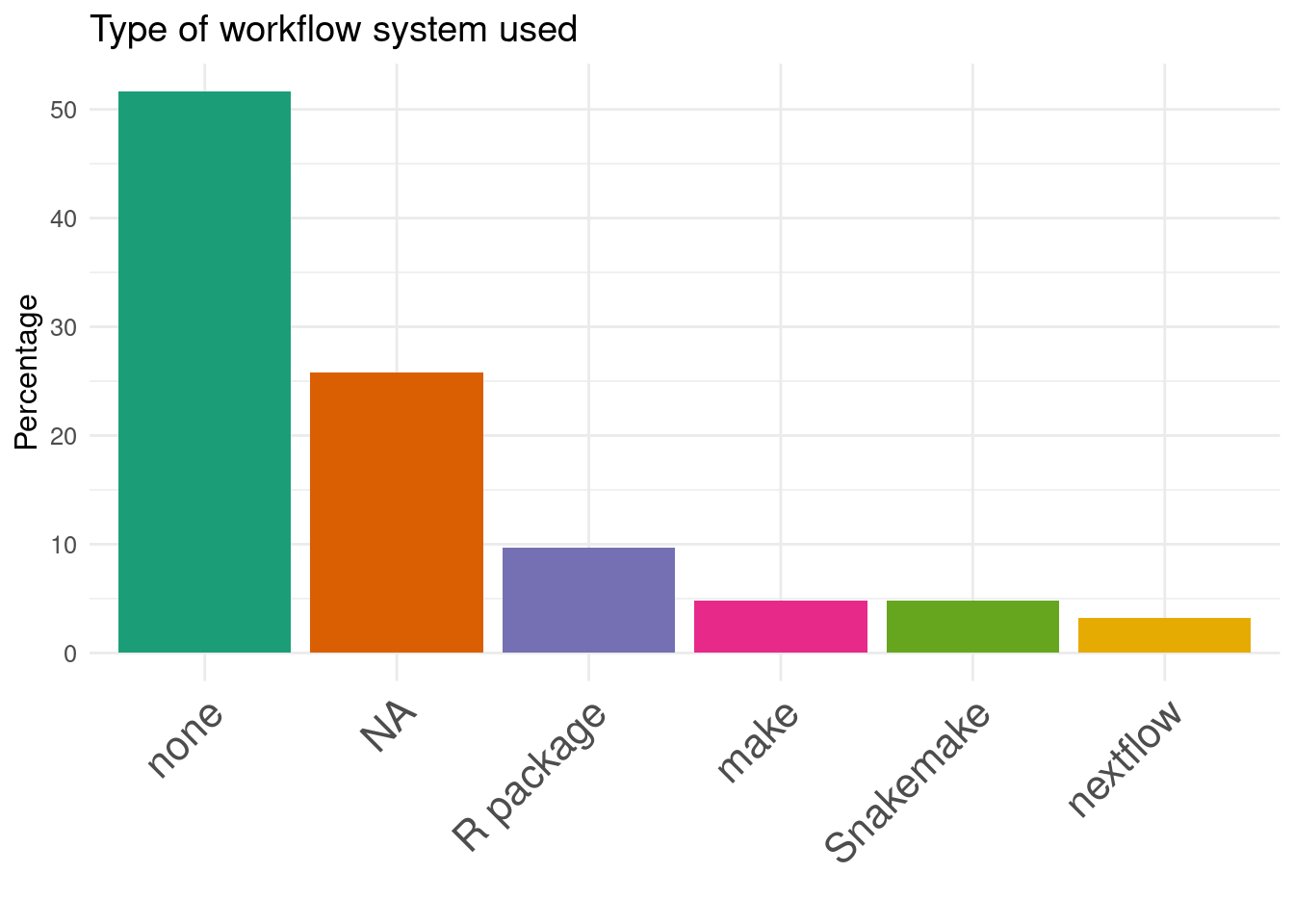
Benchmark code is available but not extensible
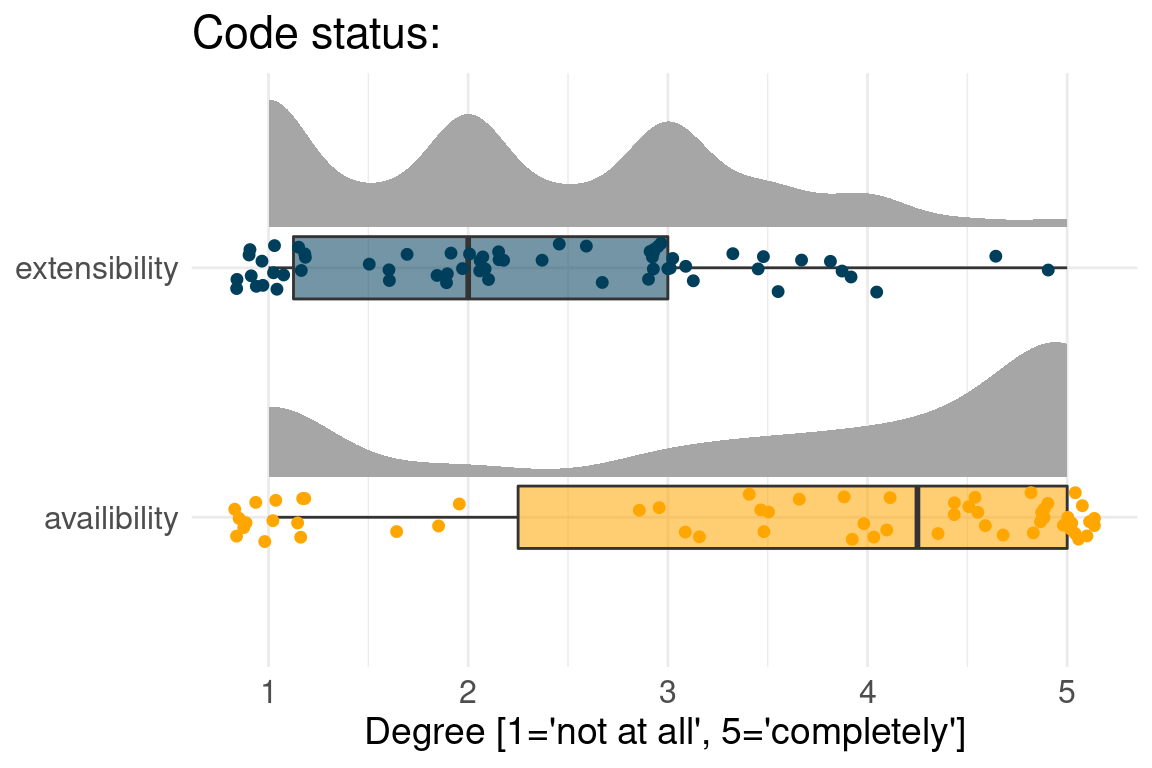
Raw input data are available, but not results
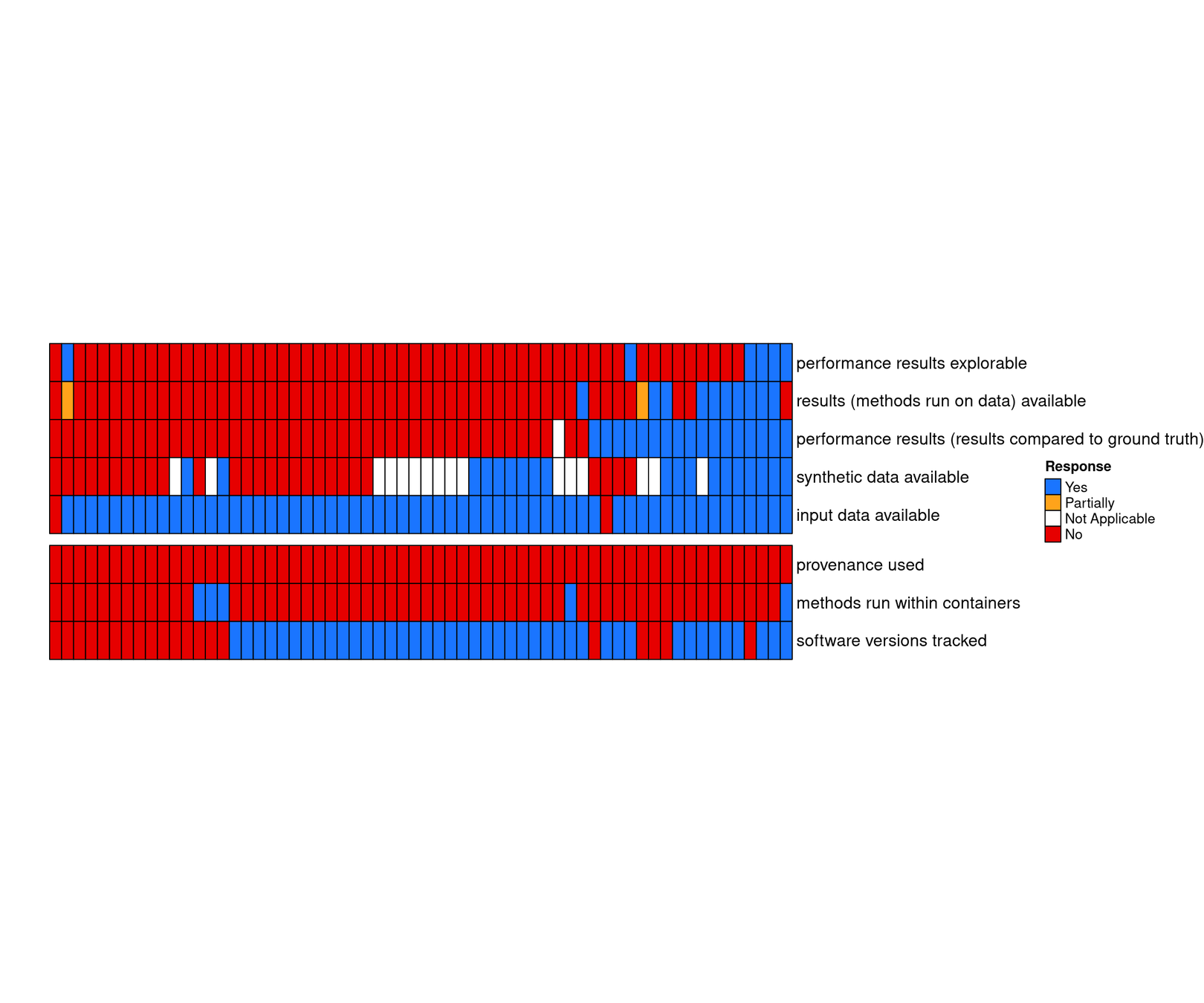
Open and continuous benchmarking
Code
available
extensible
reusable

currently part of most benchmarks
not part of current standards
Open and continuous benchmarking
Data
inputs
simulations
results

Code
available
extensible
reusable

currently part of most benchmarks
not part of current standards
Open and continuous benchmarking
Data
inputs
simulations
results

Reproducibility
versions
environments
workflows

Code
available
extensible
reusable

currently part of most benchmarks
not part of current standards
Open and continuous benchmarking
Data
inputs
simulations
results

Reproducibility
versions
environments
workflows

scale
comprehensive
continuous
Code
available
extensible
reusable

currently part of most benchmarks
not part of current standards
Omnibenchmark:
open, continuous and collaborative benchmarking




A platform for collaborative and continuous benchmarking
Method developer/
Benchmarker
Method user
Methods
Datasets



Metrics
Omnibenchmark
Goals:
- continuous
- software environments
- workflows
- all "products" can be accessed
- anyone can contribute
Design: Benchmark modules
Data
standardized datasets


= 1 "module" (renku project )



Design: Benchmark modules
Data
standardized datasets


= 1 "module" (renku project )



Methods
method results

Design: Benchmark modules
Data
standardized datasets


= 1 "module" (renku project )



Methods
method results

Metrics
metric results

Design: Benchmark modules
Data
standardized datasets


= 1 "module" (renku project )



Methods
method results

Metrics
metric results

Dashboard
interactive result exploration
Design: Benchmark modules
Data
standardized datasets


= 1 "module" (renku project )



Methods
method results

Metrics
metric results

Dashboard
interactive result exploration
Method user
Method developer/
Benchmarker
modules are connected through data bundles
Data X

Data y

Data Z



= 1 "data bundle" (data files + meta data)
= 1 "module" (renku project )



modules are connected through data bundles
Data X

Data y

Data Z



= 1 "data bundle" (data files + meta data)
= 1 "module" (renku project )






modules are connected through data bundles
Data X

Data y

Data Z



= 1 "data bundle" (data files + meta data)
= 1 "module" (renku project )






process

modules are connected through data bundles
Data X

Data y

Data Z



= 1 "data bundle" (data files + meta data)
= 1 "module" (renku project )






process

Method 1

Method 2


modules are connected through data bundles
Data X

Data y

Data Z



= 1 "data bundle" (data files + meta data)
= 1 "module" (renku project )






process

Method 1

Method 2


Method 3

Benchmark runs are coordinated by an Orchestrator
Data X

Data y

Data Z

process




Method 1

Method 2

Method 3



Orchestrator
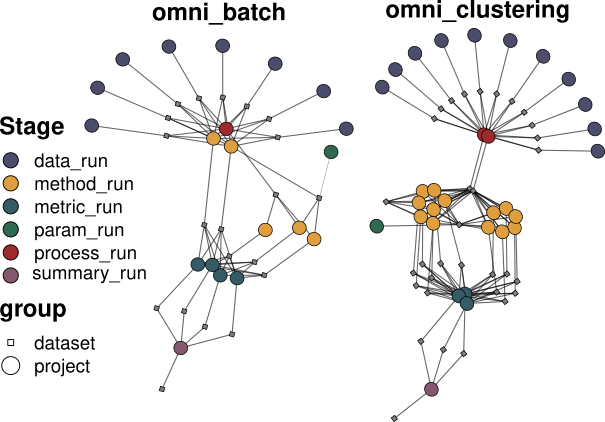
omnibenchmark allows flexible architectures
bettr: A better way to explore what is best
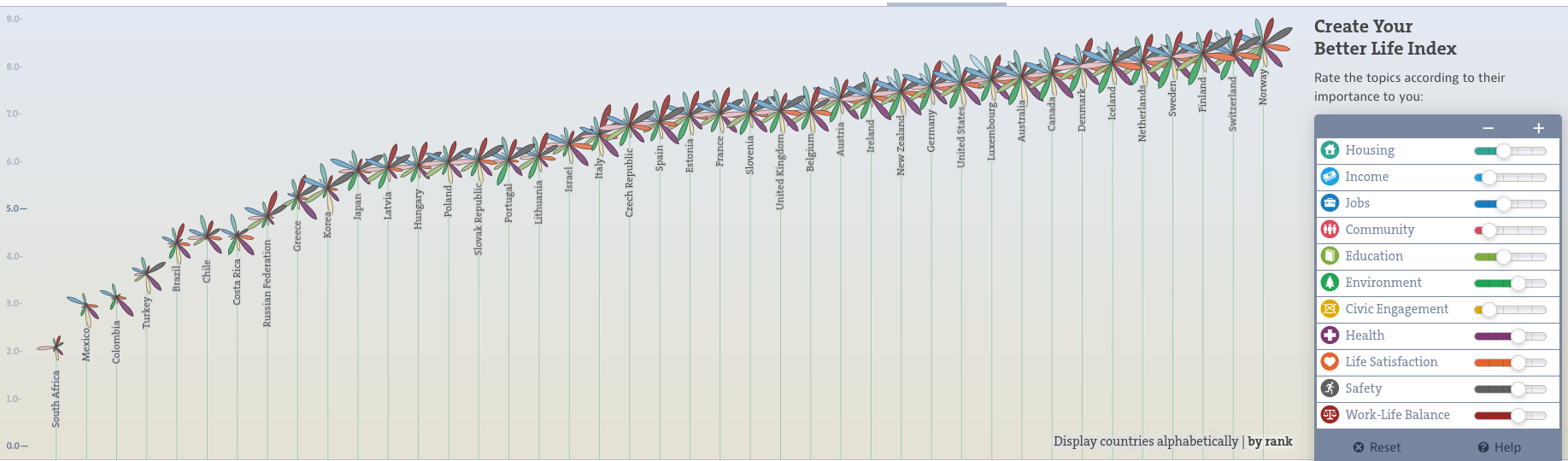
https://www.oecdbetterlifeindex.org
bettr: A better way to explore what is best
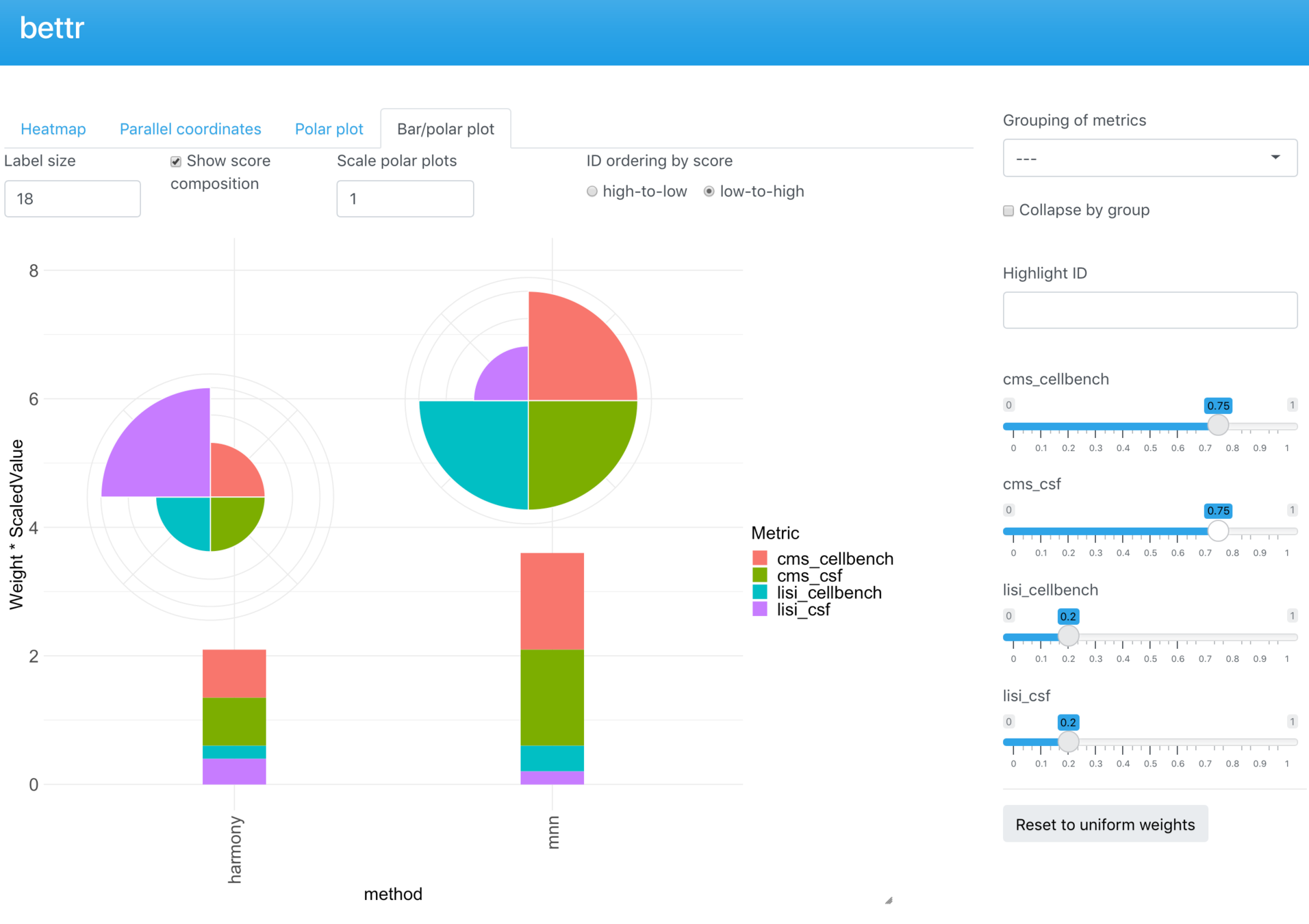
Omnibenchmark
Data

Reproducibility

scale
Code

part of omnibenchmark
not part of omnibenchmark
Omnibenchmark
Data

Reproducibility

scale
Code
available
extensible
reusable

part of omnibenchmark
not part of omnibenchmark
Omnibenchmark
Data
inputs
simulations
results

Reproducibility

scale
Code
available
extensible
reusable

part of omnibenchmark
not part of omnibenchmark
Omnibenchmark
Data
inputs
simulations
results

Reproducibility
versions
environments
workflows

scale
Code
available
extensible
reusable

part of omnibenchmark
not part of omnibenchmark
Omnibenchmark
Data
inputs
simulations
results

Reproducibility
versions
environments
workflows

scale
(comprehensive)
continuous
comparable
Code
available
extensible
reusable

part of omnibenchmark
not part of omnibenchmark
(Open) questions?
(Open) questions?
- Technical maturity for open and continuous benchmarking?
easy
flexible
(Open) questions?
- Technical maturity for open and continuous benchmarking?
- What about authorities and risks for biases?
easy
flexible
(Open) questions?
- Technical maturity for open and continuous benchmarking?
- Are we asking the "right" questions?
- What about authorities and risks for biases?
easy
flexible
(Open) questions?
- Technical maturity for open and continuous benchmarking?
- Are we asking the "right" questions?
- What about authorities and risks for biases?
- Who pays and for how long?
easy
flexible

Acknowledgement

omnibenchmark: current status
Against the ’one method fits all data sets’ philosophy
- there is not one way to benchmark different methods
- the same benchmark can provide different/multiple conclusions
--> Dynamic, extensible and explorable benchmarking system
Collaborative benchmarking: METHOD DEVELOPER
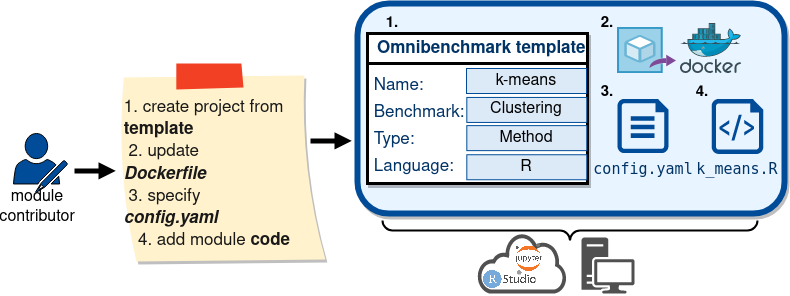
Collaborative benchmarking: bENCHMARKER
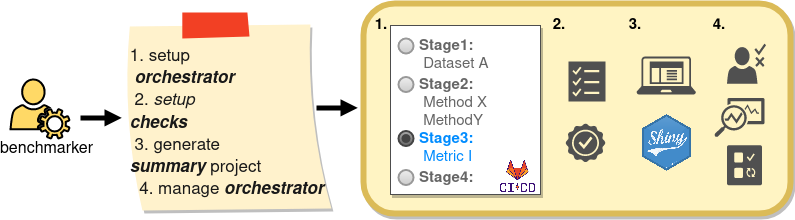
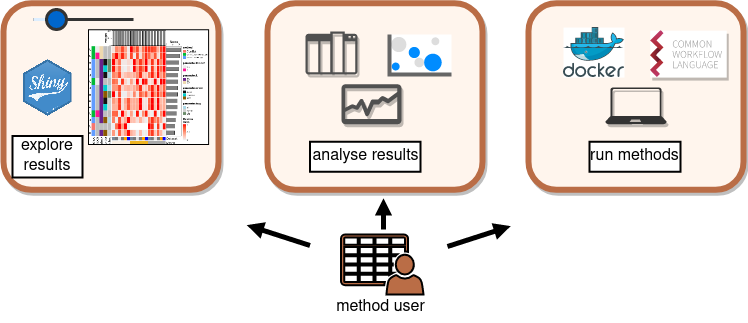
Collaborative benchmarking: mETHOD uSER
the knowledge graph: benchmark metadata are stored as triples

Result
Code
generated
used_by
has_attribute
keyword
has_attribute
keyword
Data
Code
Result
used_by
generated
User interaction with renku client
Automatic triplet generation
Triplet store "Knowledge graph"
User interaction with renku client
KG-endpoint queries
Omnibenchmark components
contributer
user

omnibenchmark-python

omniValidator
benchmarker
projects

templates

omb-site

{
orchestrator

triplestore


omni-sparql
dashboards

Orchestrator coordinate benchmark runs

GitLab

Docker

Workflow

Module:
Template code
Module code


Data bundle
GitLFS/S3
Input/Output files
Metadata

Workflow manager are rarely used
Batch effects: example
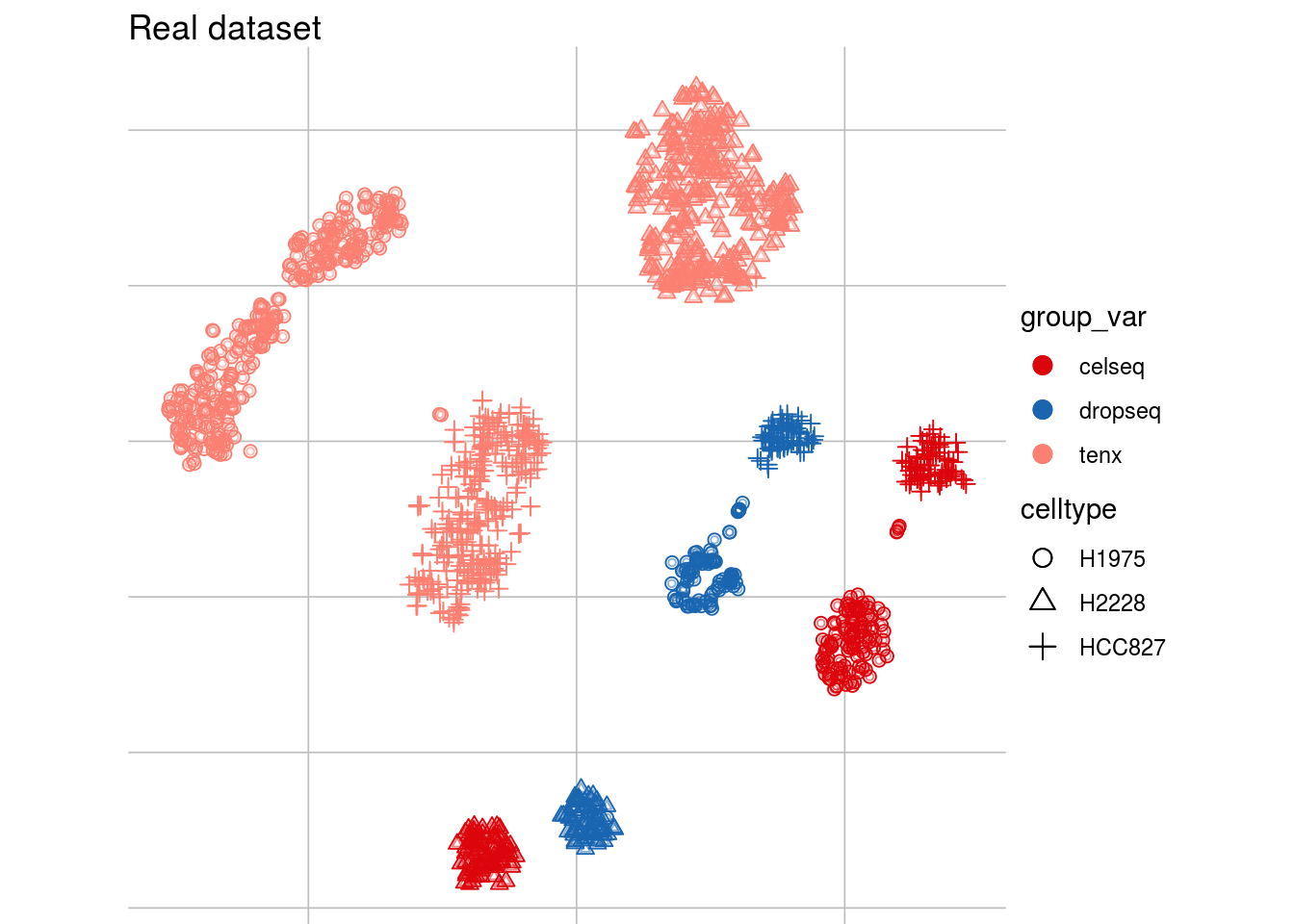
tsne1
tsne2
Batch effects: Why do we care?
Batch effects: Why do we care?
Batch effects: characteristics
Metrics vary in their batch detection ranges

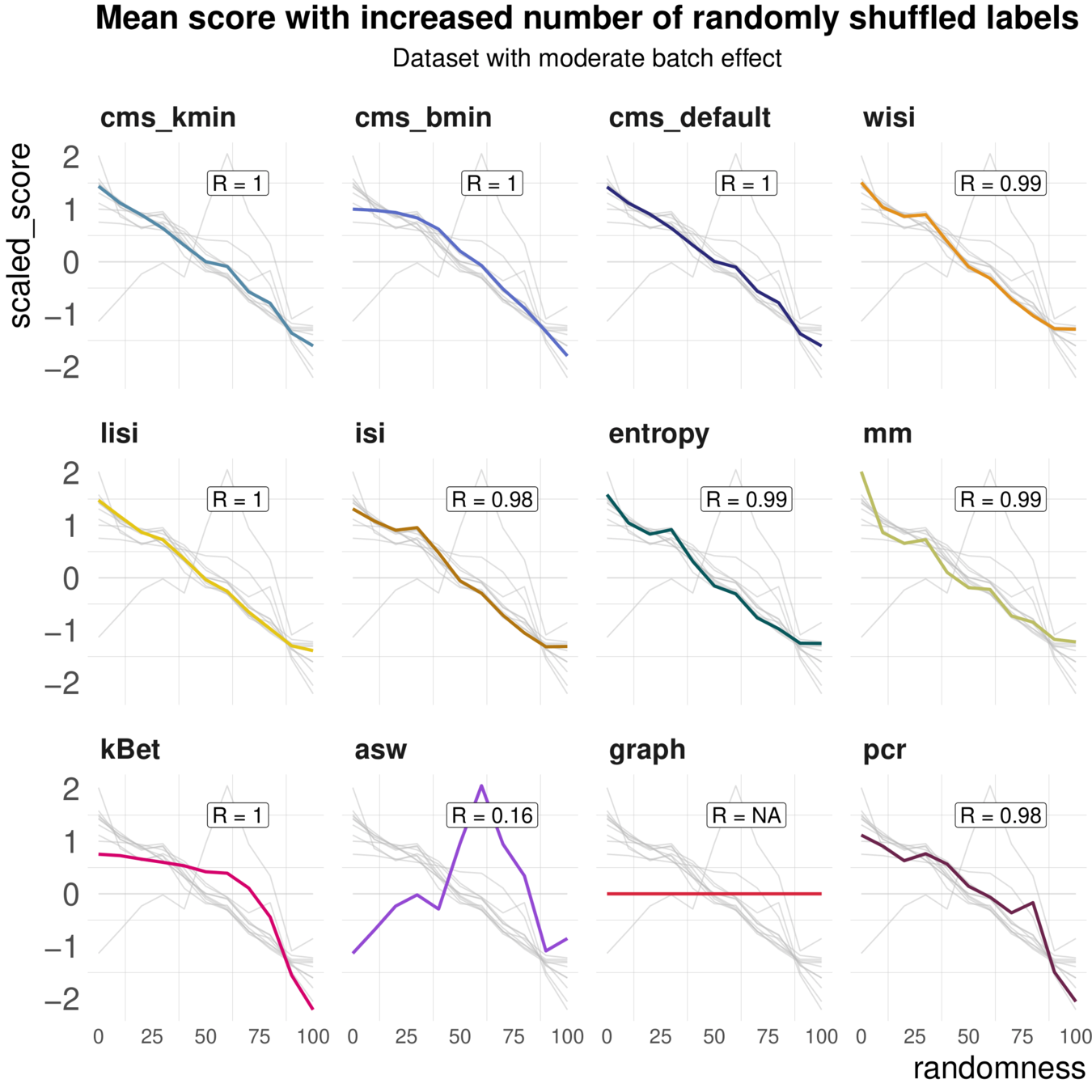
Task 2: Batch label permutation
Aim: Negative control and test whether metrics scale with randomness
Spearman correlation of metrics with the percentage of randomly permuted batch label
Task 2: increasing randomness
Most metrics scale with increasing randomness
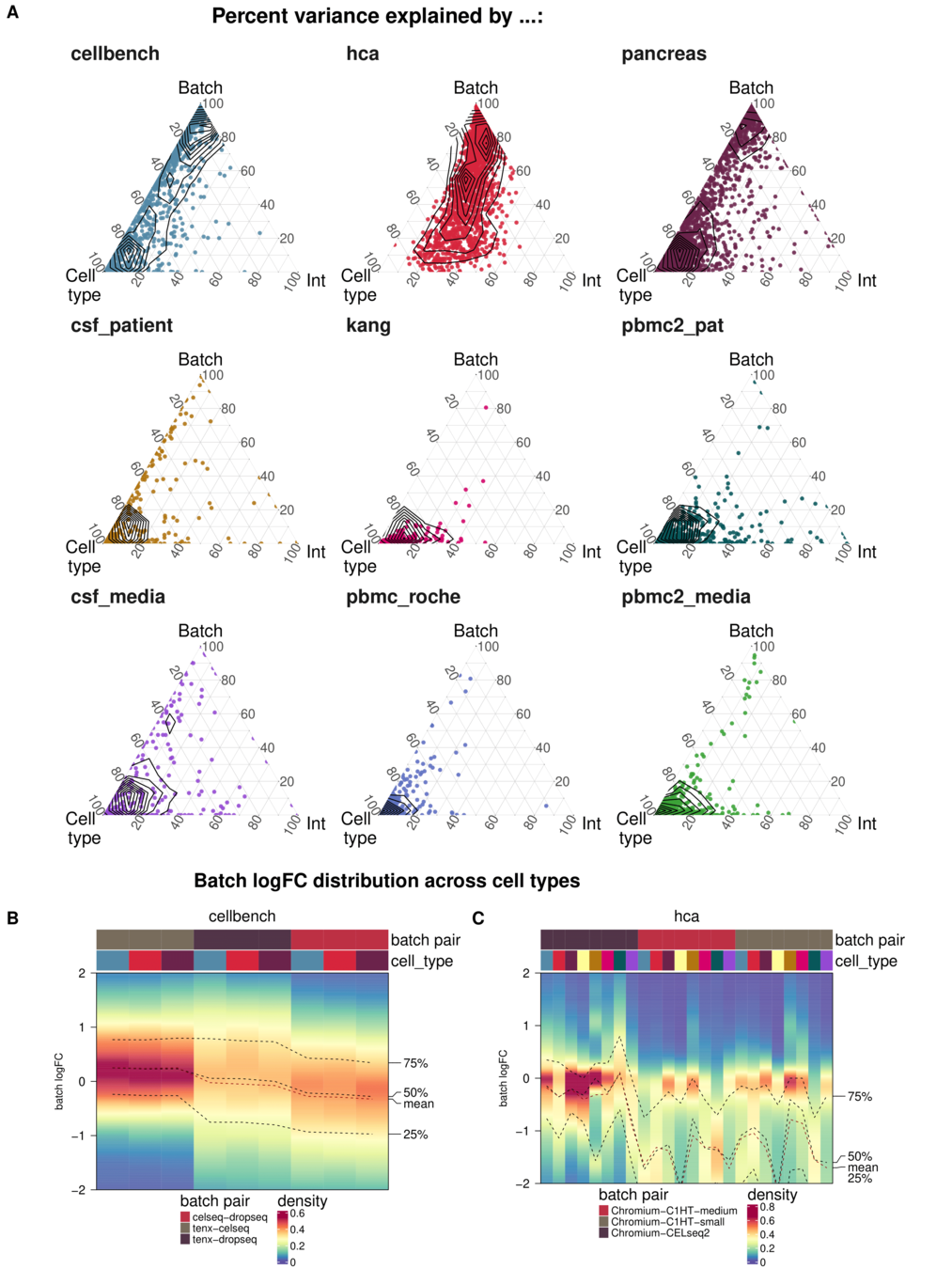
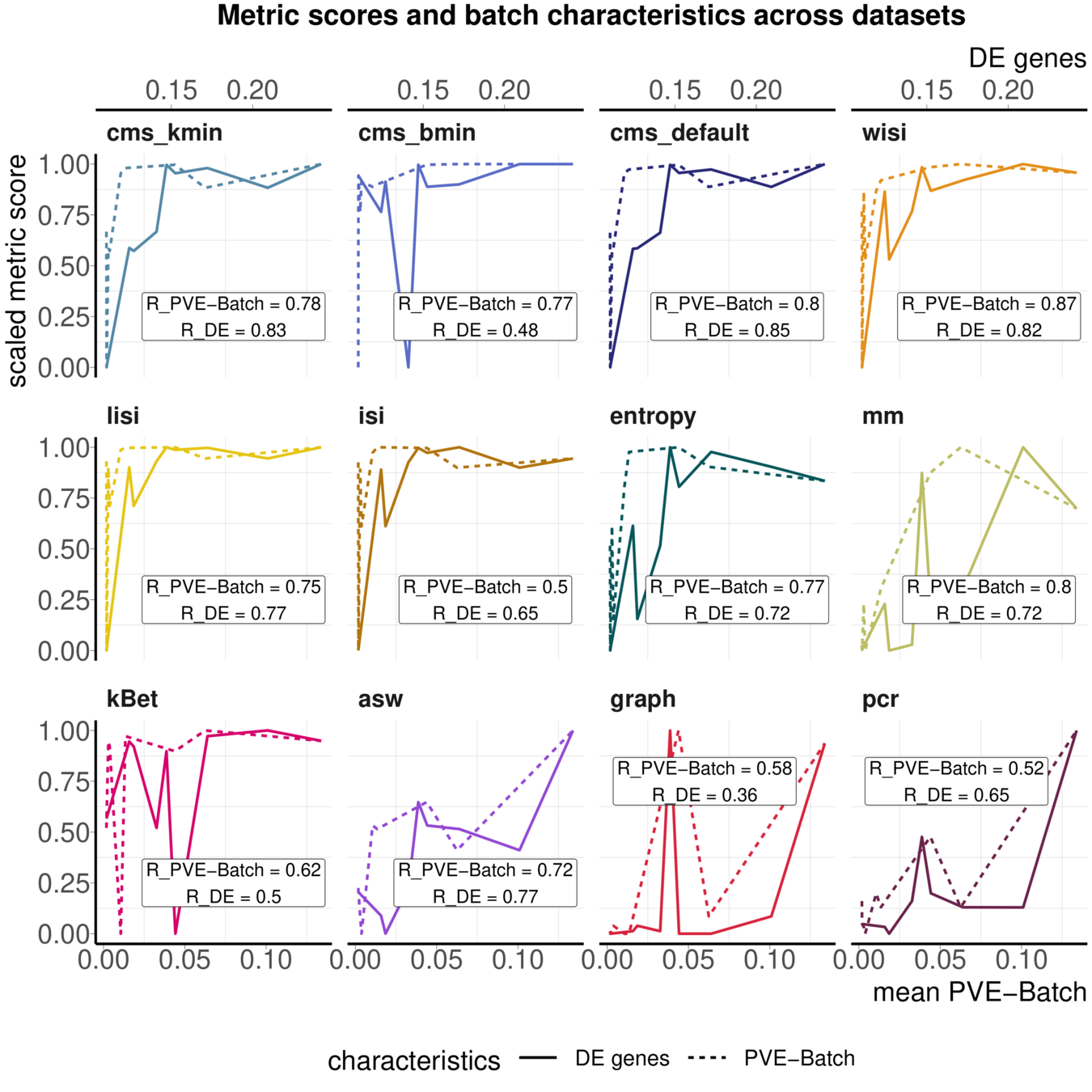
Task 3: Batch characteristics
Aim: Test whether metrics reflect batch strength across datasets
Spearman correlation of metrics with surrogates of batch strength (e.g., percent variance explained by batch (PVE-Batch) and proportion of DE genes between batches) across datasets
Task 3: batch characteristics


Variance attribution
Batch DE genes
Across batch comparisons are limited
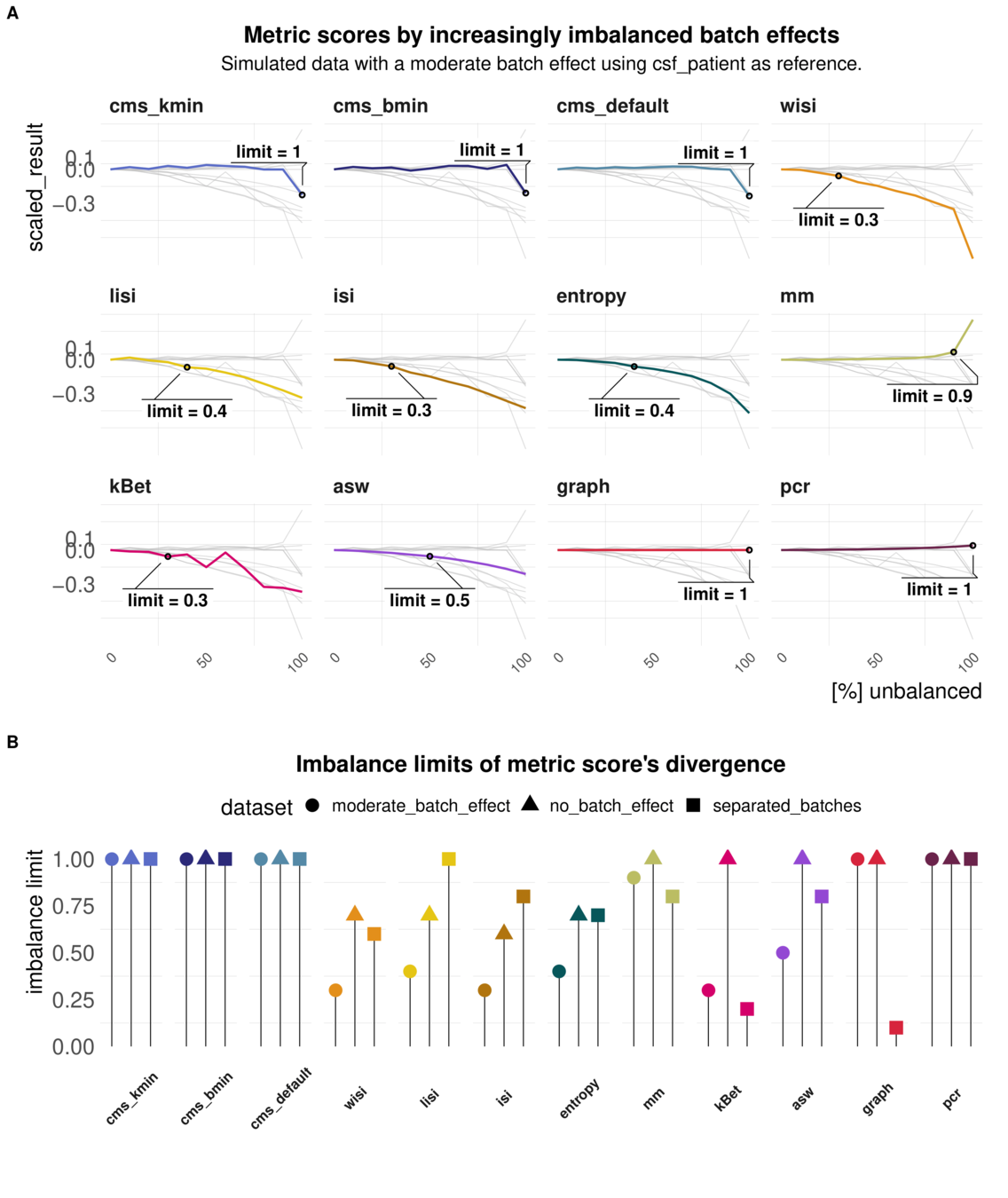
Task 4: Imbalanced batches
Aim: Reaction of metrics to imbalanced cell type abundance within the same dataset
Test sensitivity towards imbalance of cell type abundance
Task 4: Imbalanced batches
Batch strength and Imbalance can be mixed
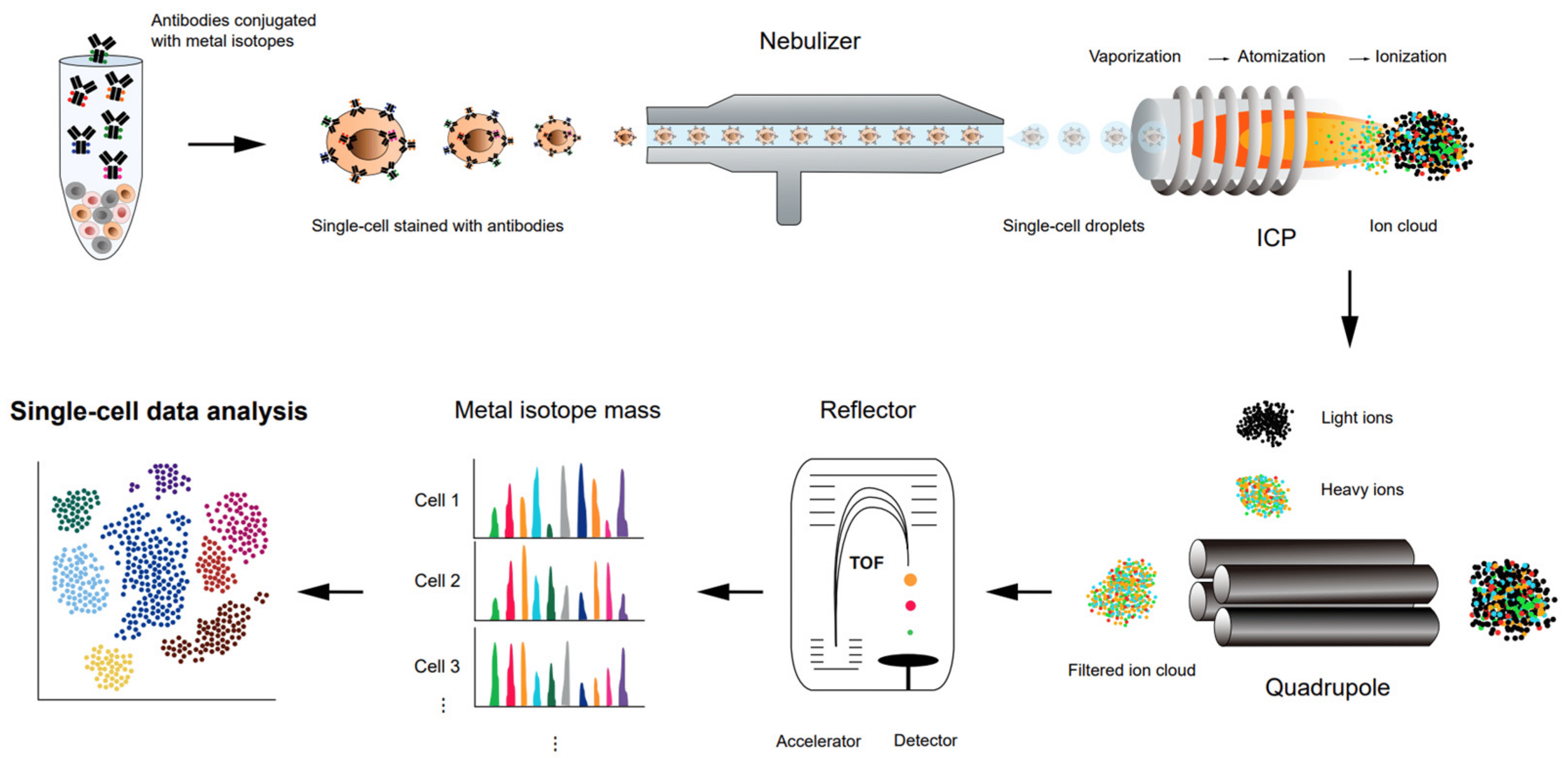
spectrometry
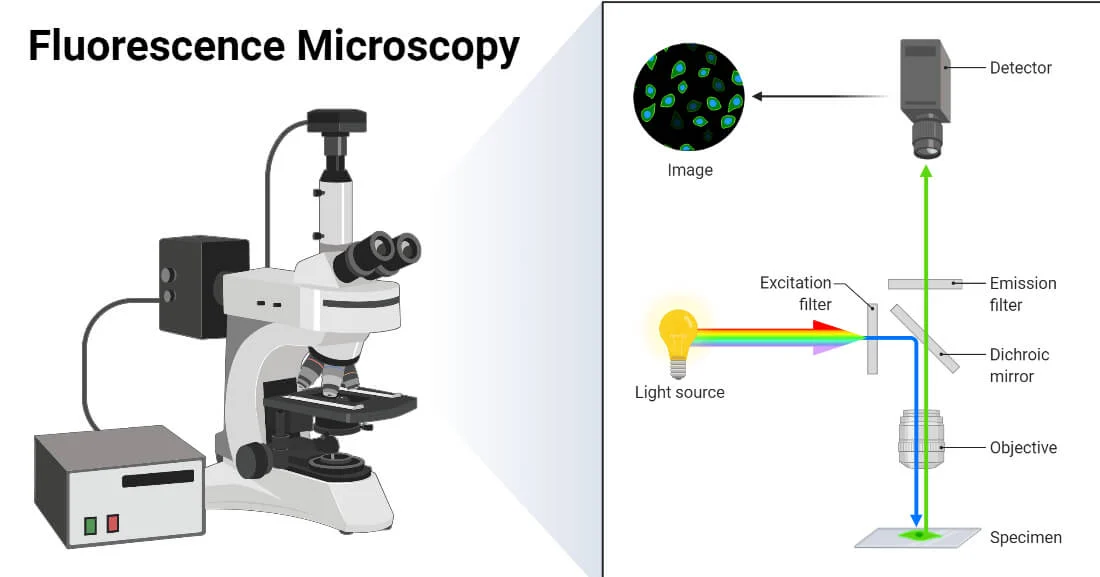
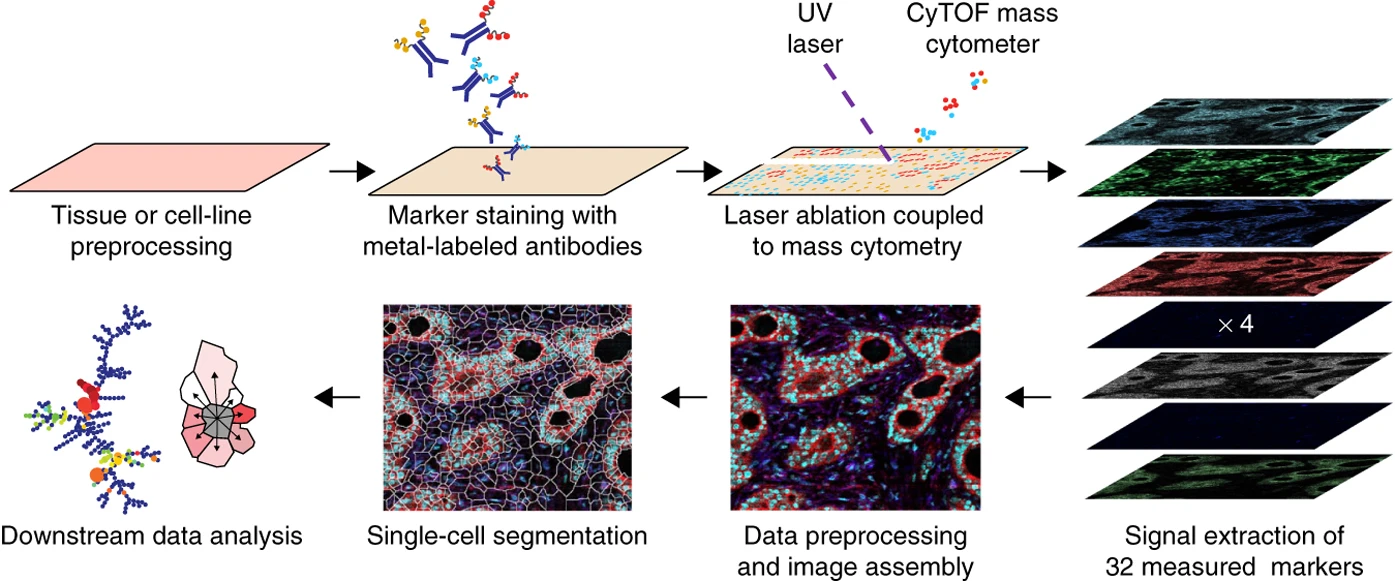
imaging
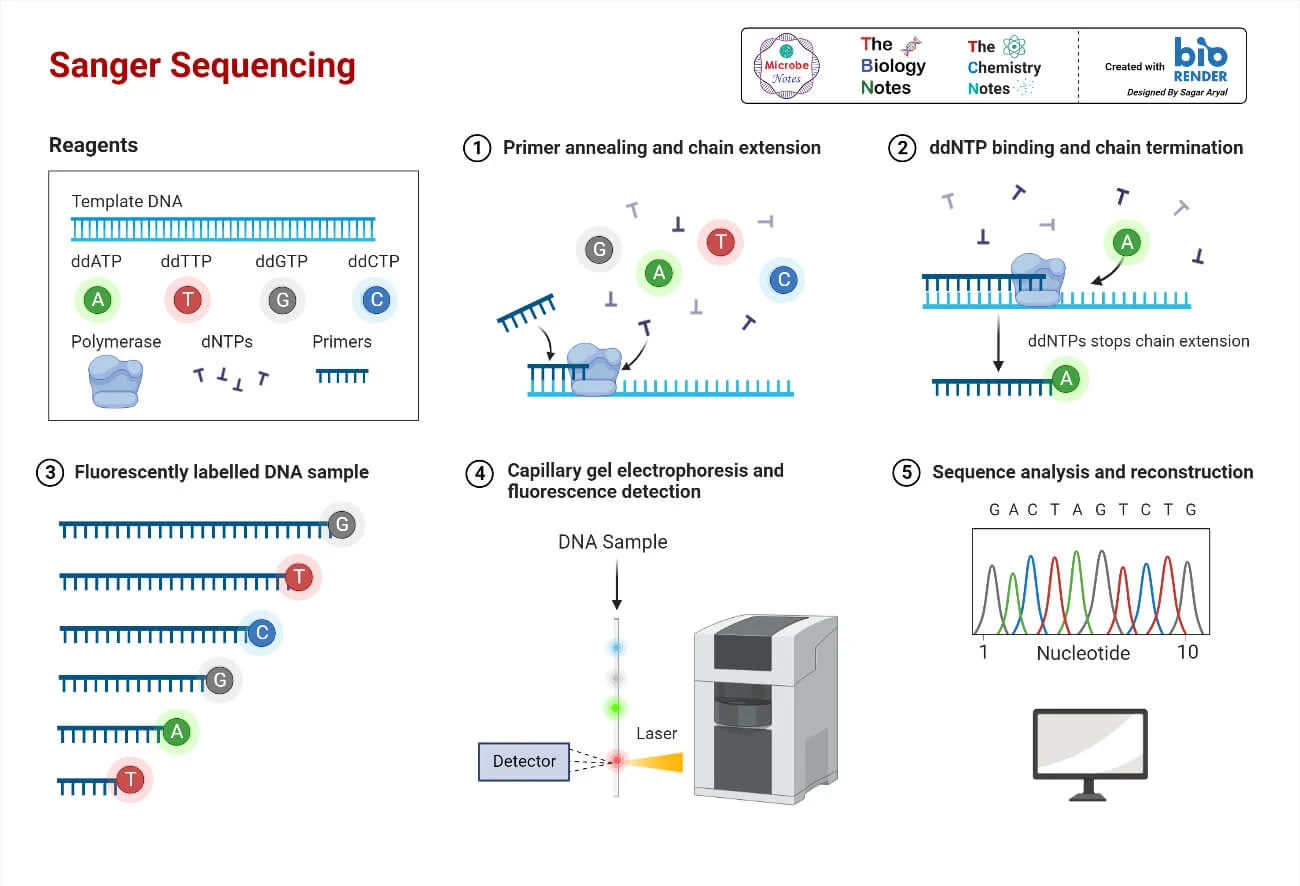

aatgctgcgctaatcgcgcgta
tcgggatcatgccctagtggcc
cgccatattggcgtcaggtcga
atcggatccggtgactccatgc
atttcaggctcactgtggcacc
sequencing
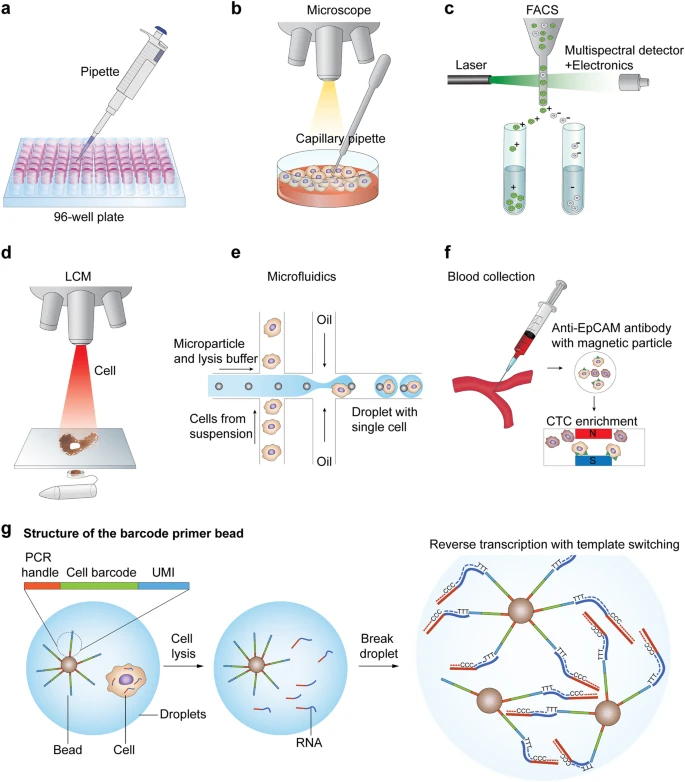
How to study Cells at molecular resolution ?











Cells are regulated at different molecular levels
Which method should I use?

Luecken et al., 2021

Defense
By Almut Luetge
Defense
- 94



