Monosynaptic Scalable Architecture Revealed by Transsynaptic Rabies Tracing
Daniel Fürth
Meletis Lab
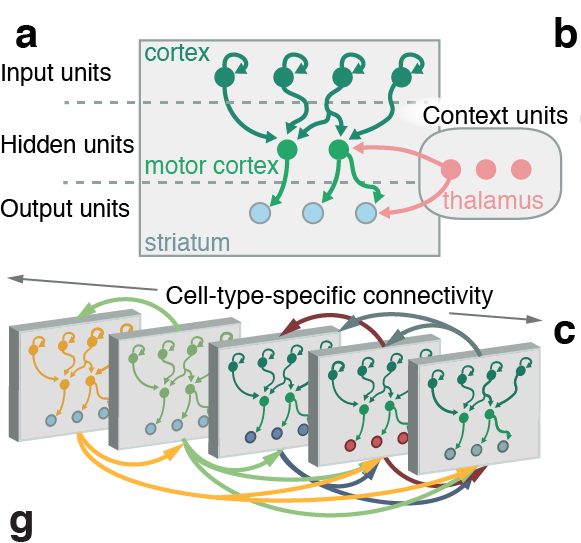
Network for Networks
12 Feb 2016
daniel.furth@ki.se
- Yang Xuan
- Ourania Tzortzi
- Iakovos Lazardis
- Konstantinos Meletis
- DMC lab
Acknowledgement

Reconstructing brain from sectioned tissue
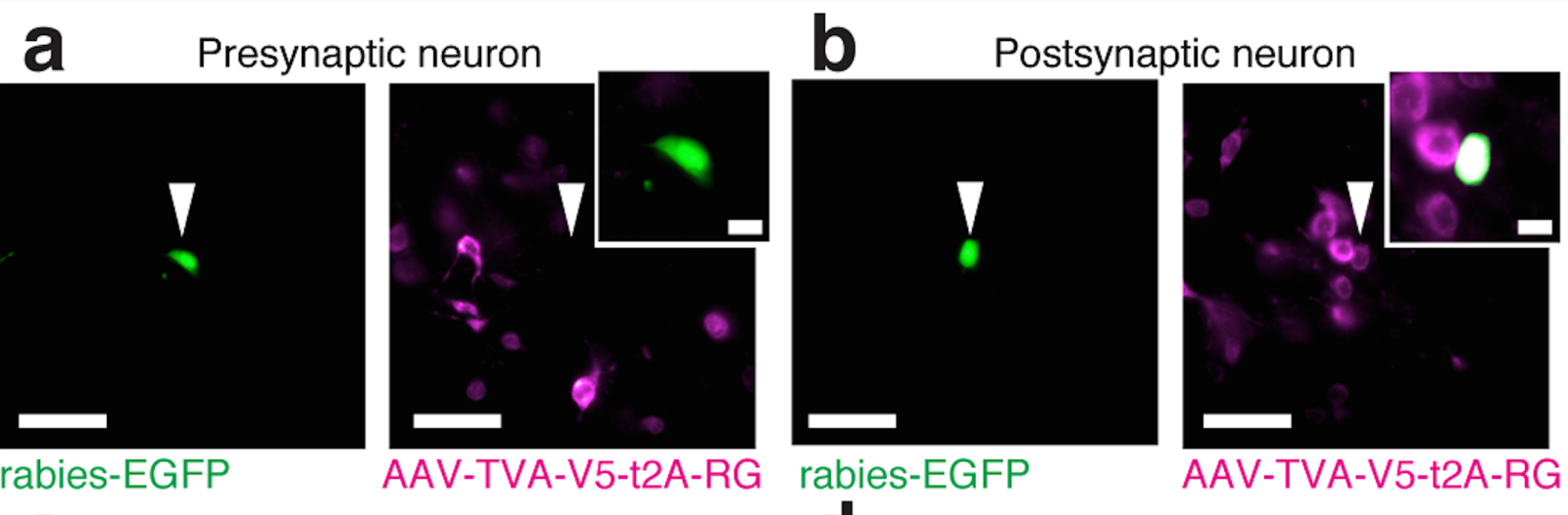
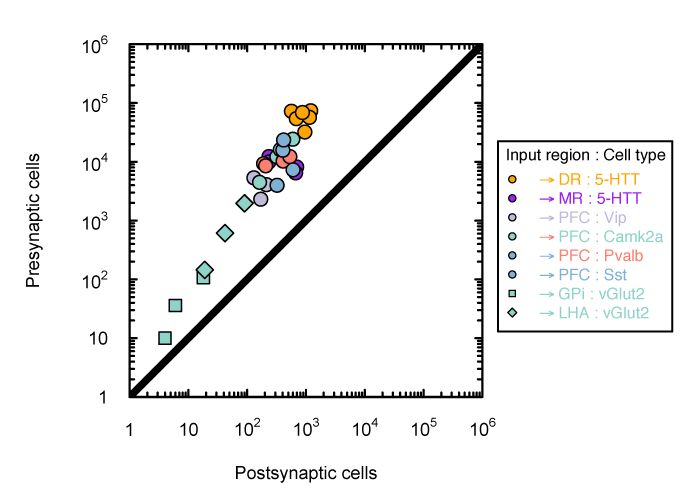
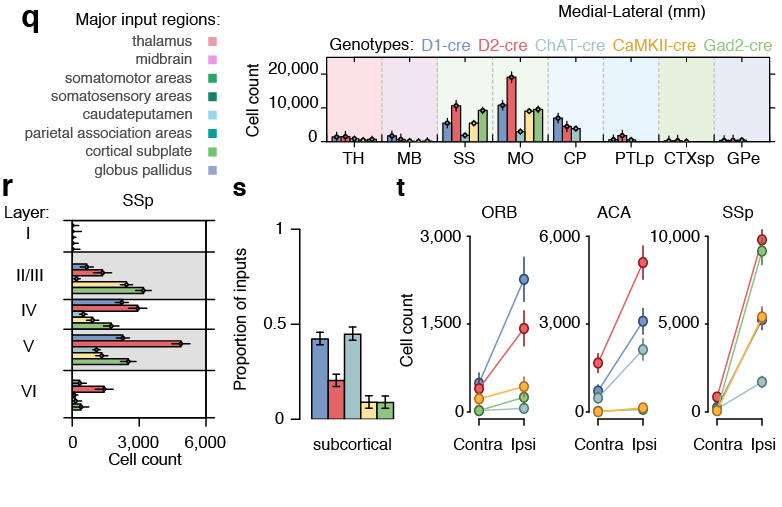
Tracing the network
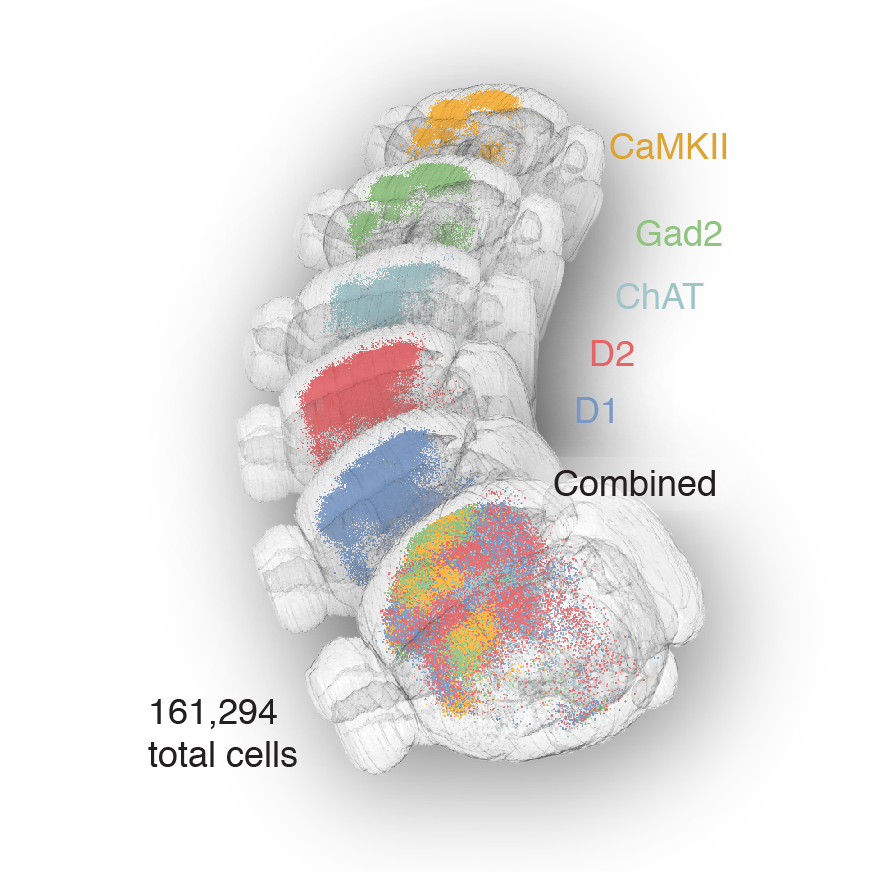
Tracing the network
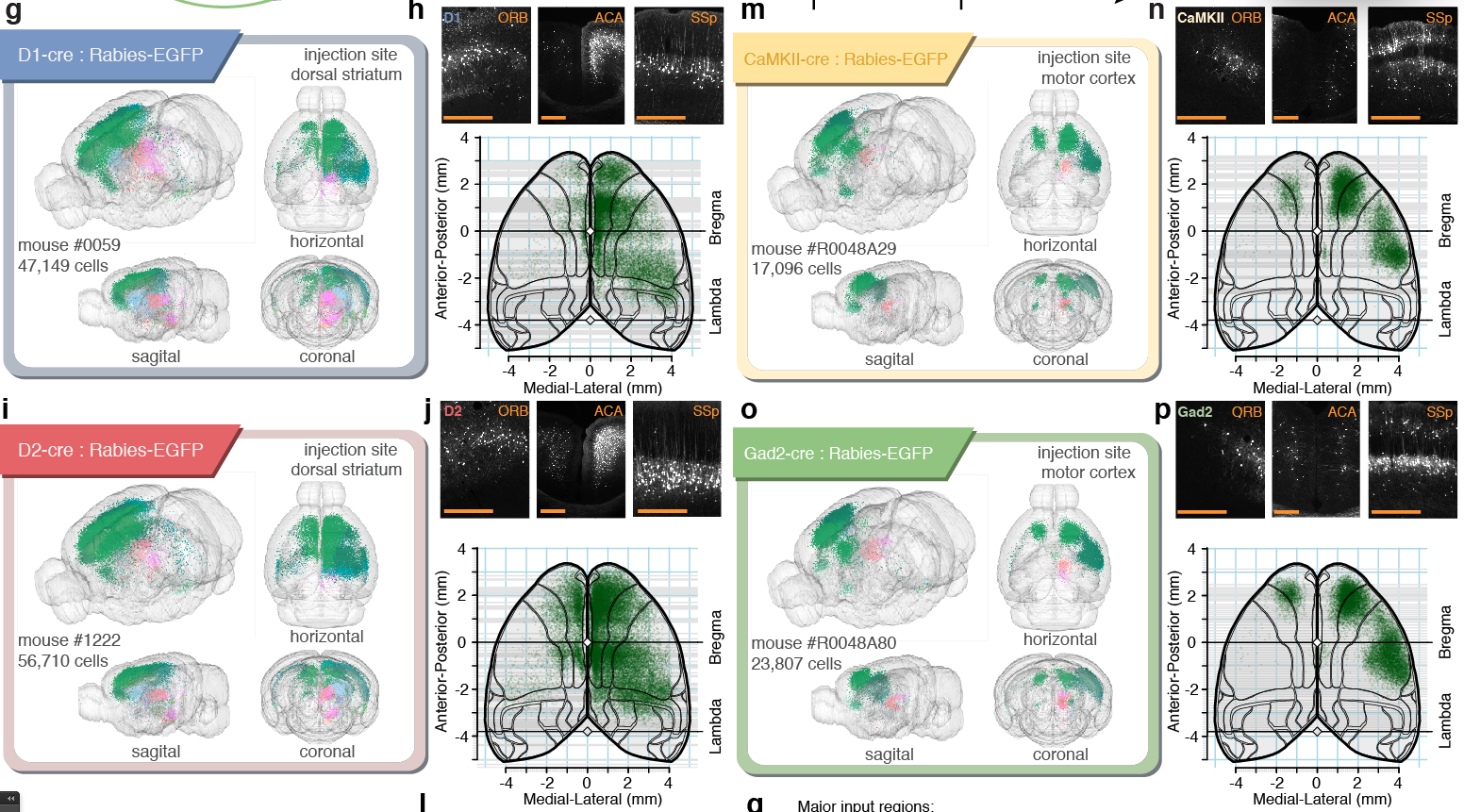
Tracing the network
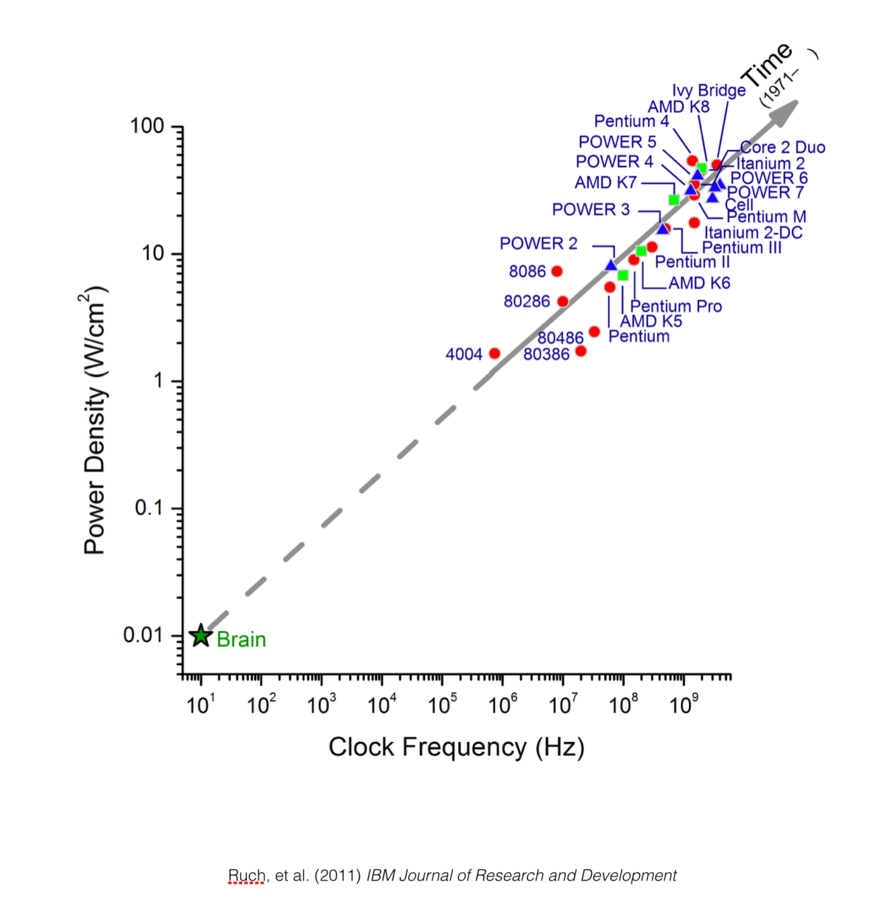
Scaling rules for rodent brians
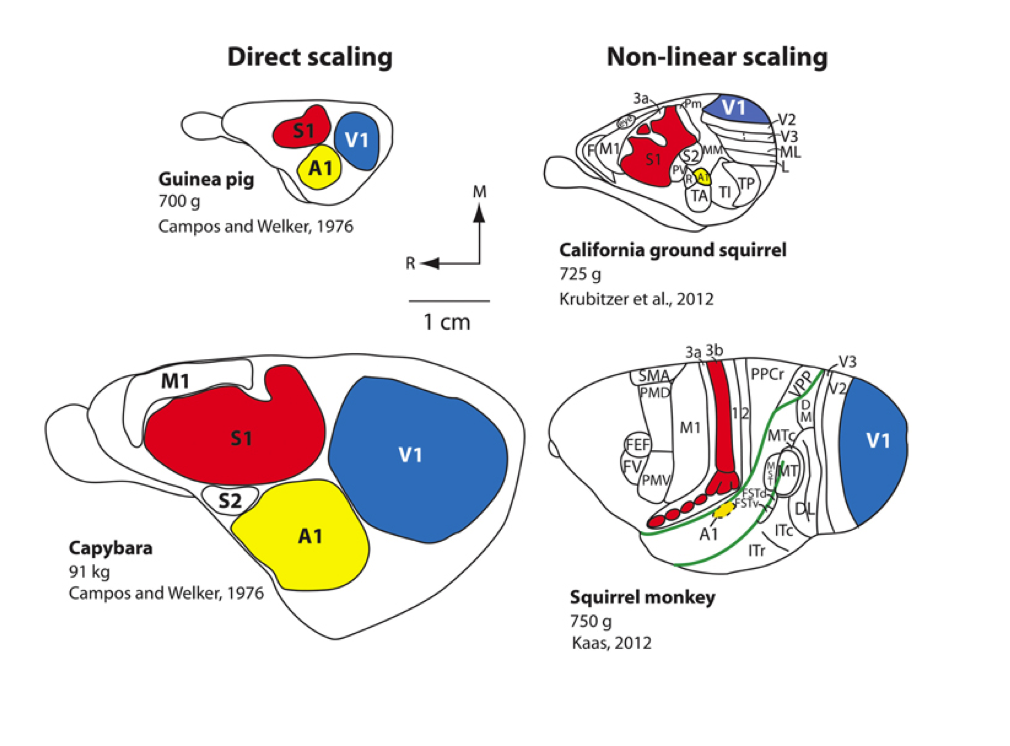
Isometric
Allometric
Cellular scaling rules for rodent brians

Herculano-Houzel et al. PNAS
Cellular scaling rules
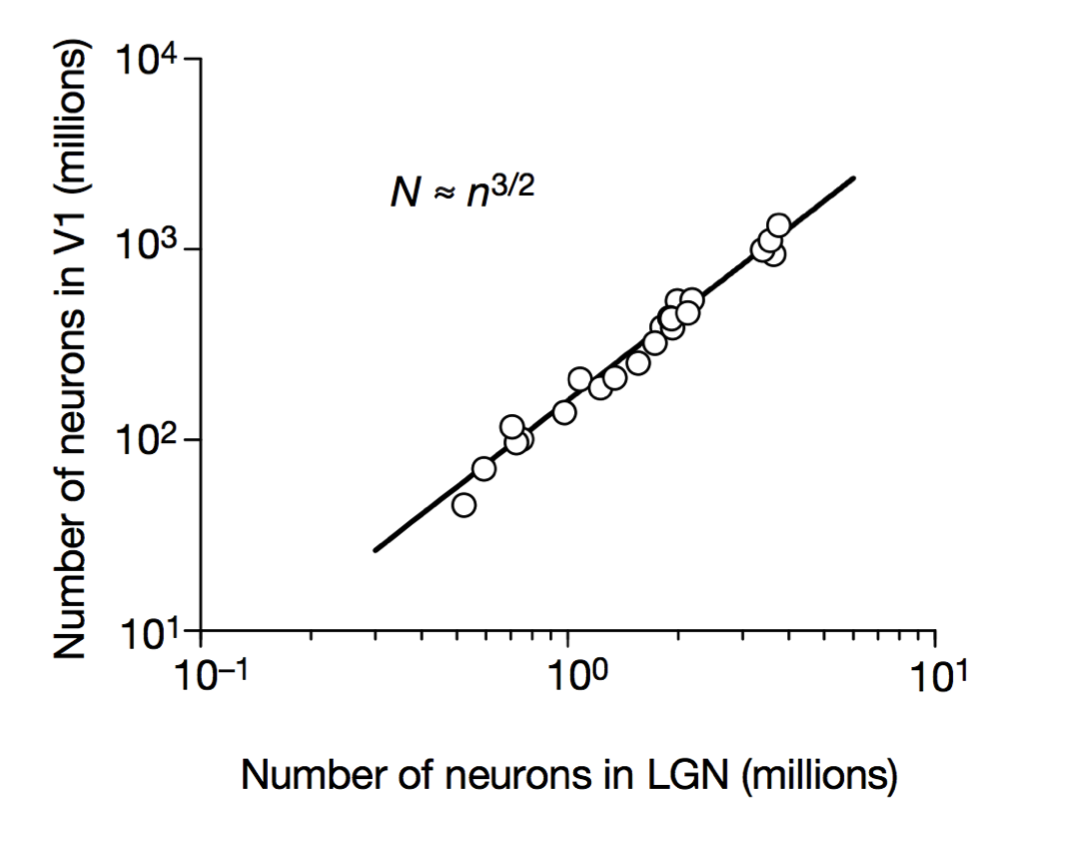
Stevens (2000) Nature
Cellular scaling rules
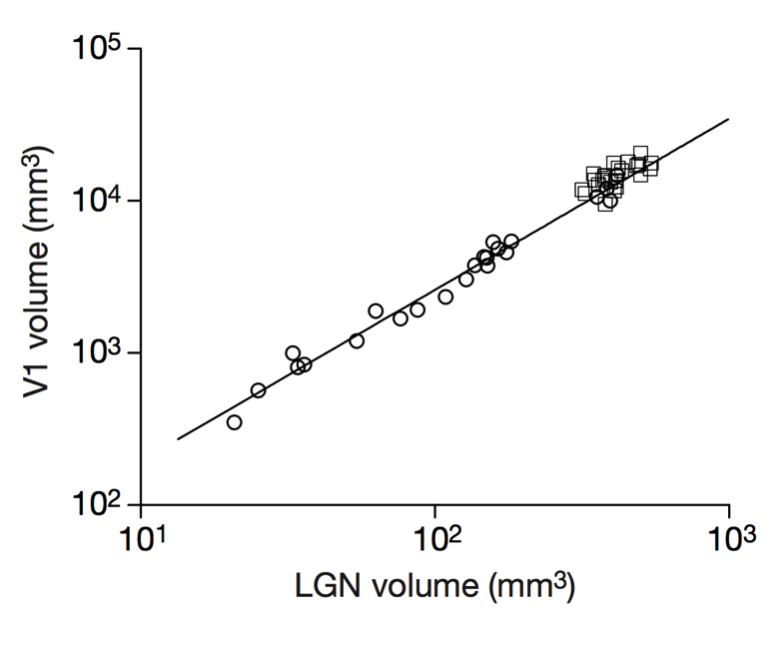
Stevens (2000) Nature
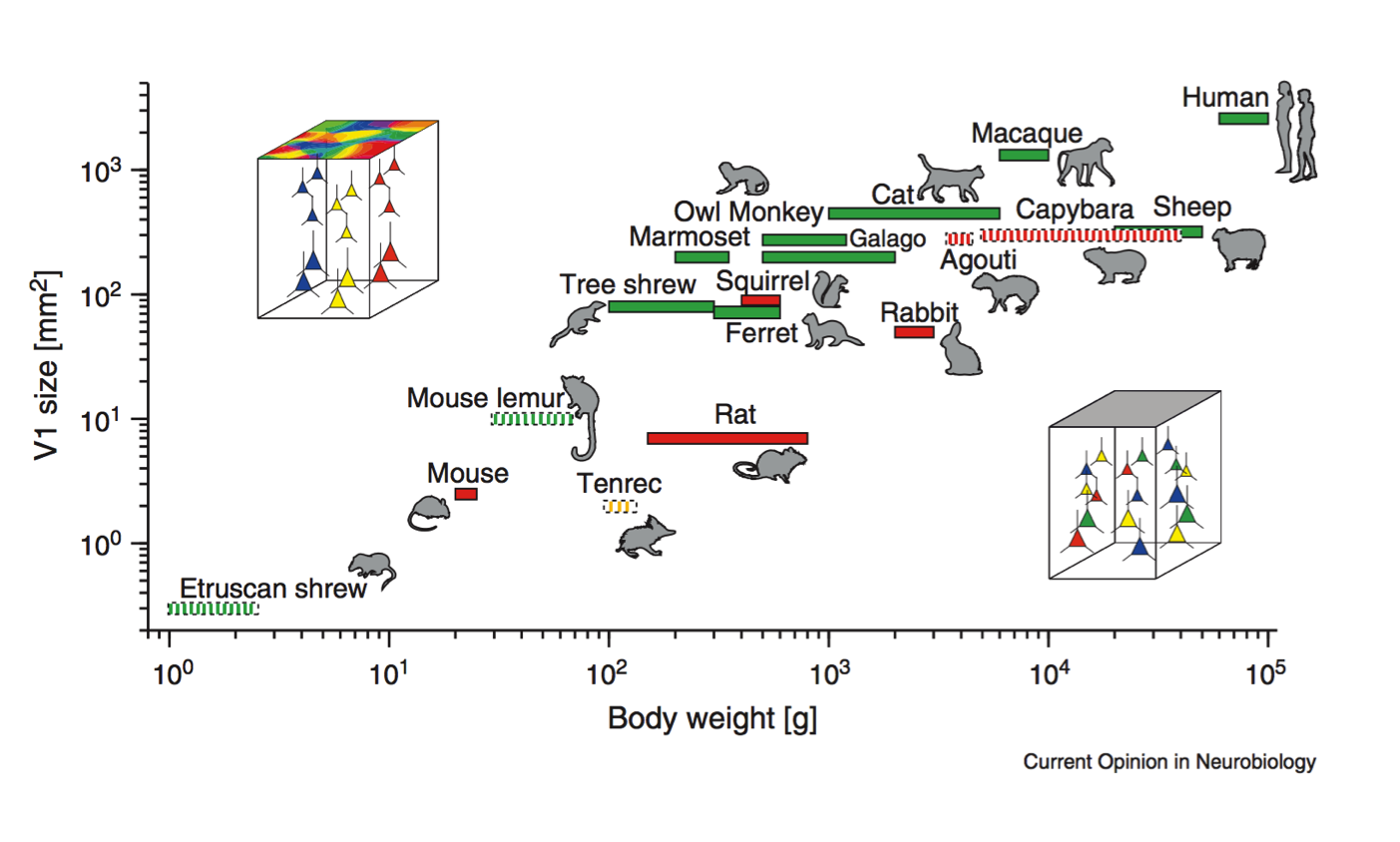
Cellular scaling rules
Hubel & Wiesel (1974), Blasdel (1986)
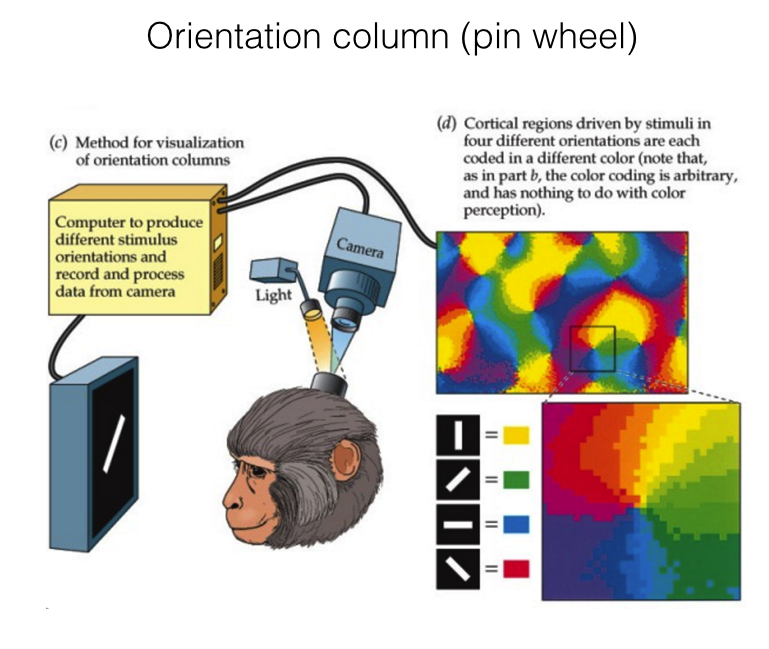
Cellular scaling rules


Cellular scaling rules
Stevens (2000) Nature
Cellular scaling rules
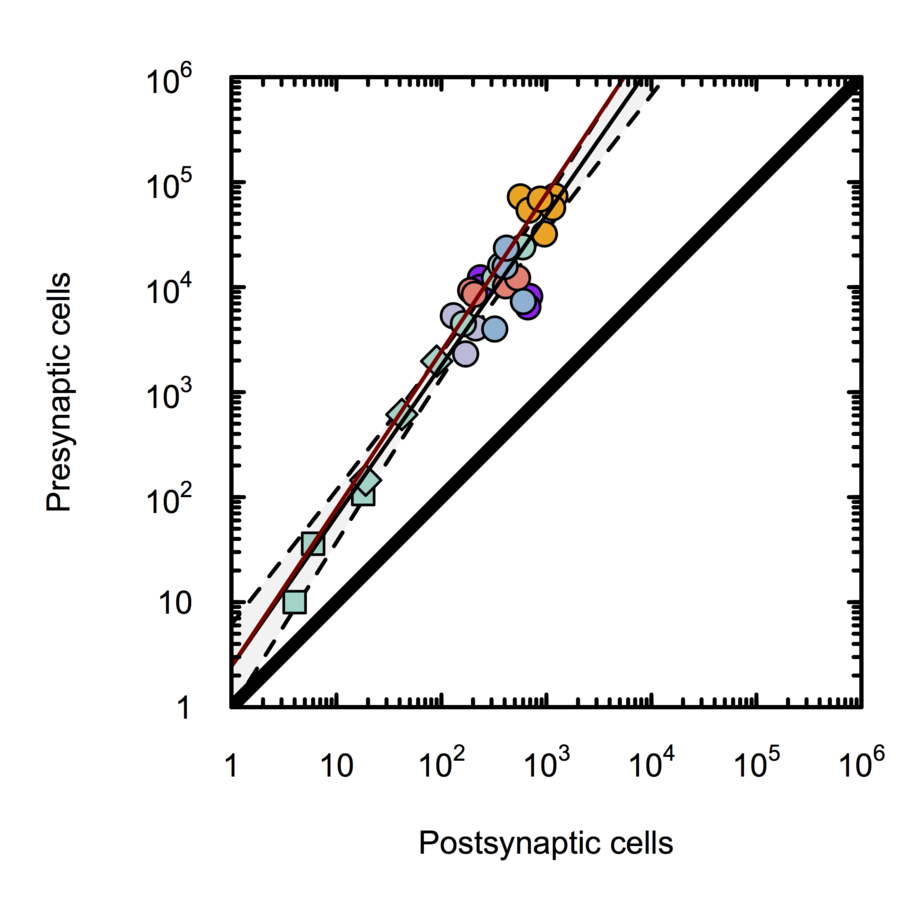
Monosynaptic scaling in mice
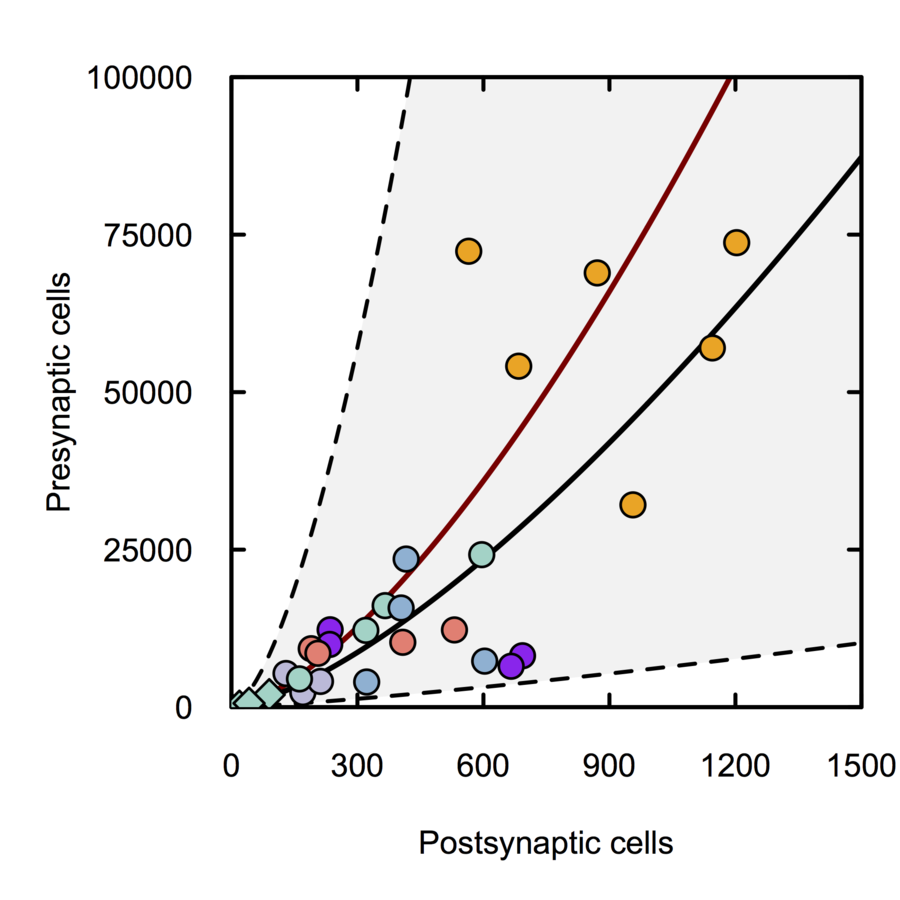
Synapse density and dendritic branching
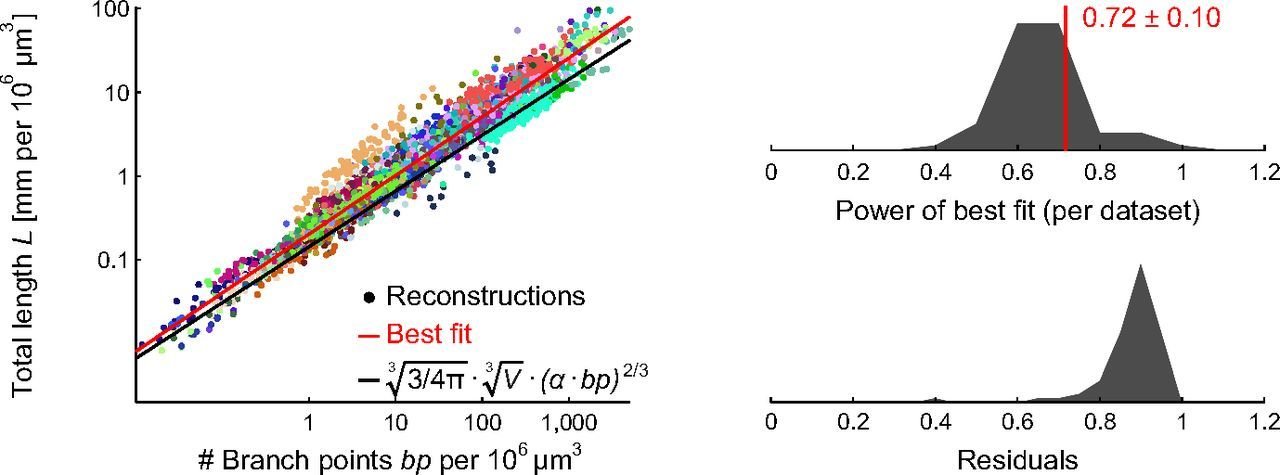
Cuntz, Mathy & Häusser (2012) PNAS
Synapse density and dendritic branching
Livneh, Feinstein, Klein and Mizrahi (2009) JoN
Synapse density and dendritic branching
Cuntz, Mathy & Häusser (2012) PNAS

Synapse density and dendritic branching
Cuntz, Mathy & Häusser (2012) PNAS
Self-organizing branching process
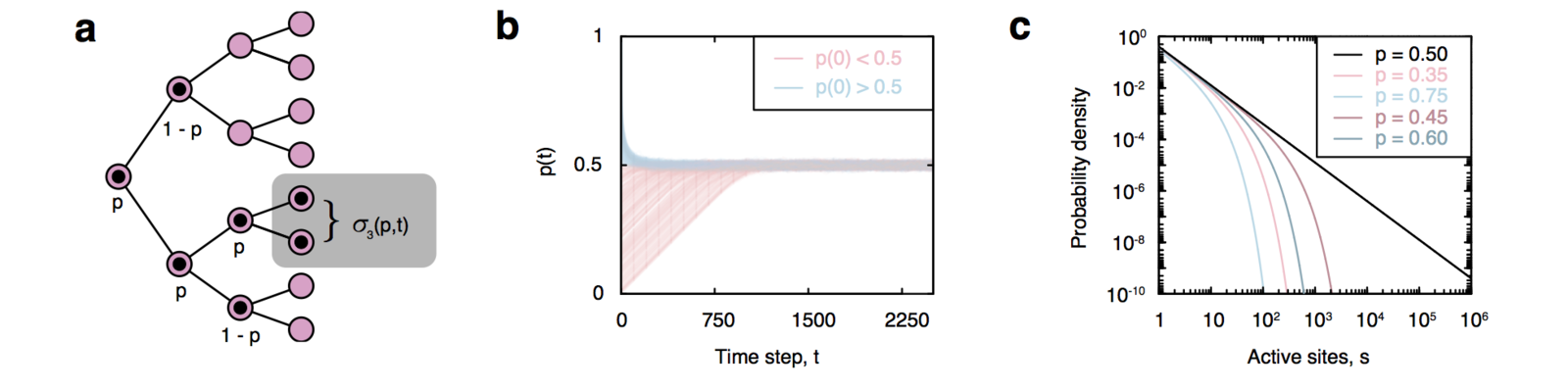
Self-organizing branching process
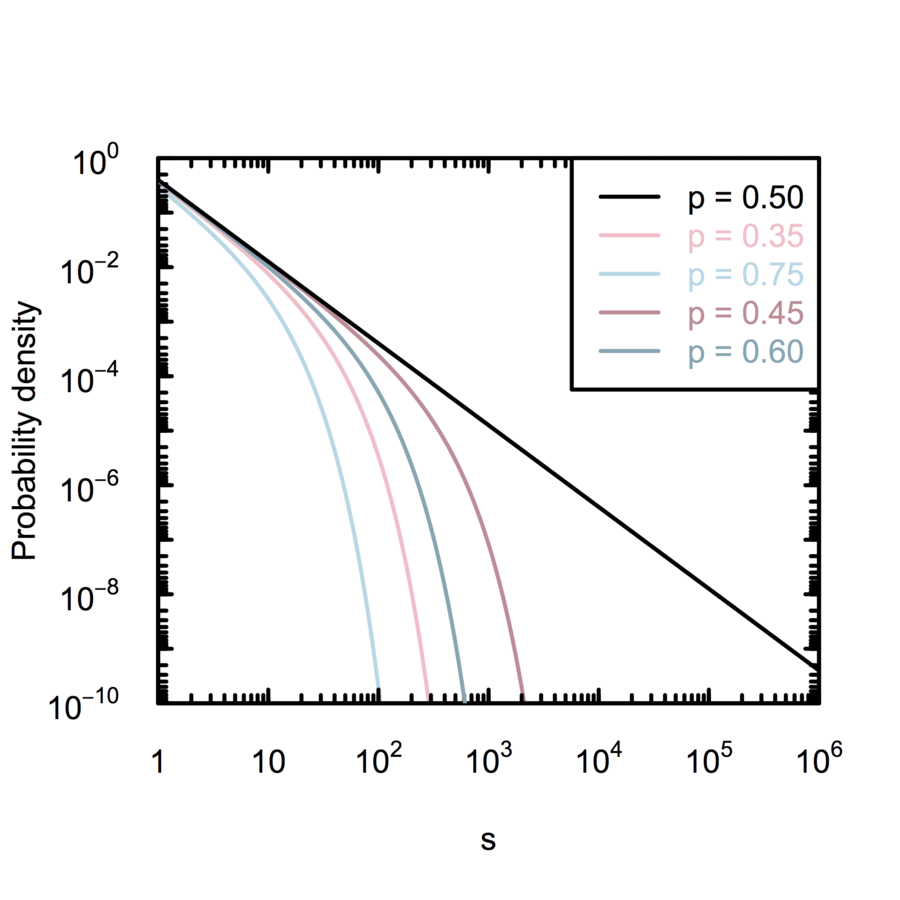
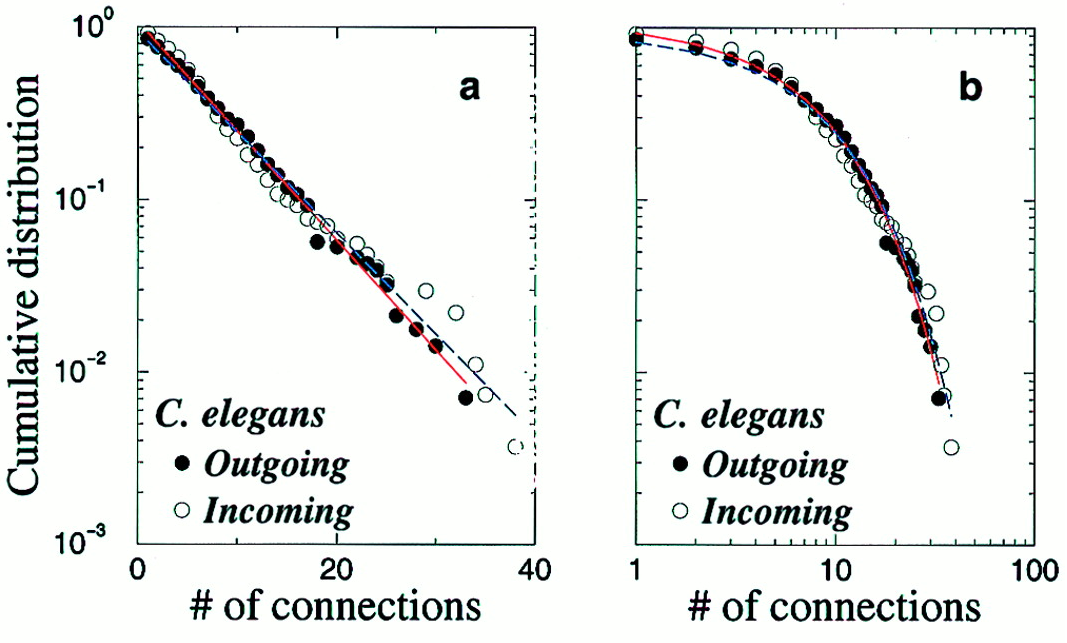
Amaral et al. (2000); White, Southgate, Thompson & Brenner (1986)
Mean-field results
Total output for the network:
Total connection of type k for ith neuron:
Average number of connections in network
Mean-field results
Cost per unit length for dendrite to cover an area
Set average number of connections to the cost for the dendritic tree
Bayesian inference
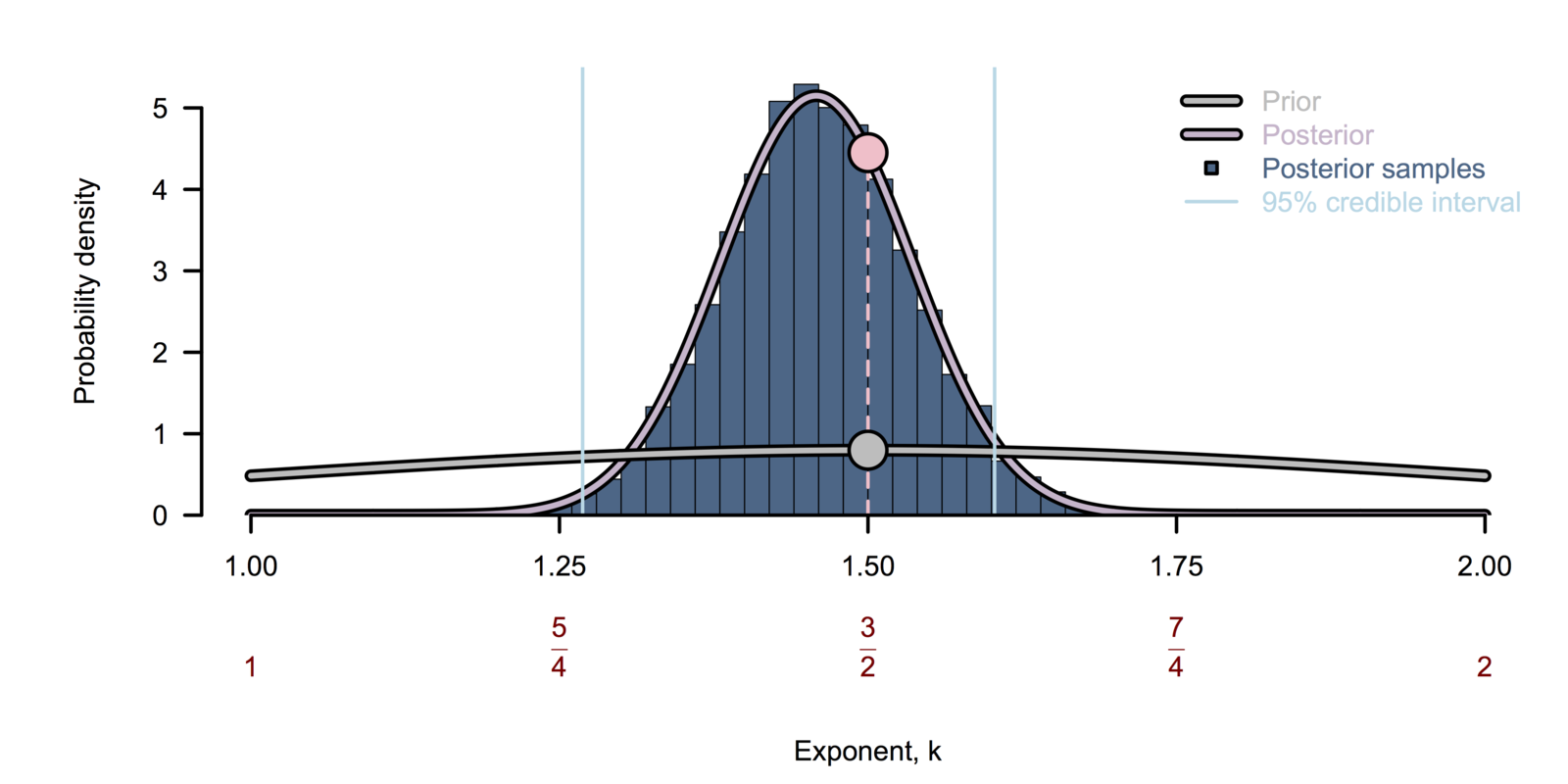
Bayesian inference
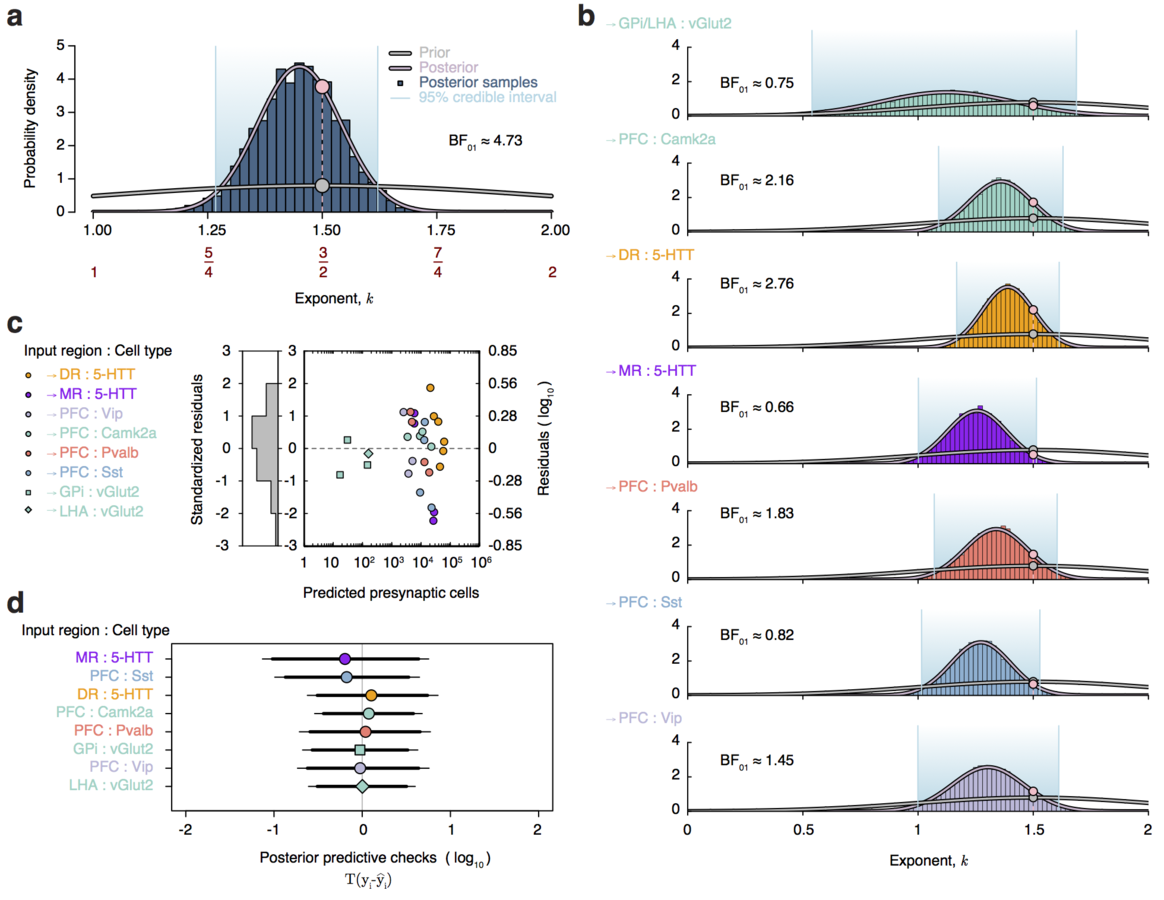
Dendritic pruning as an age-dependent branching process

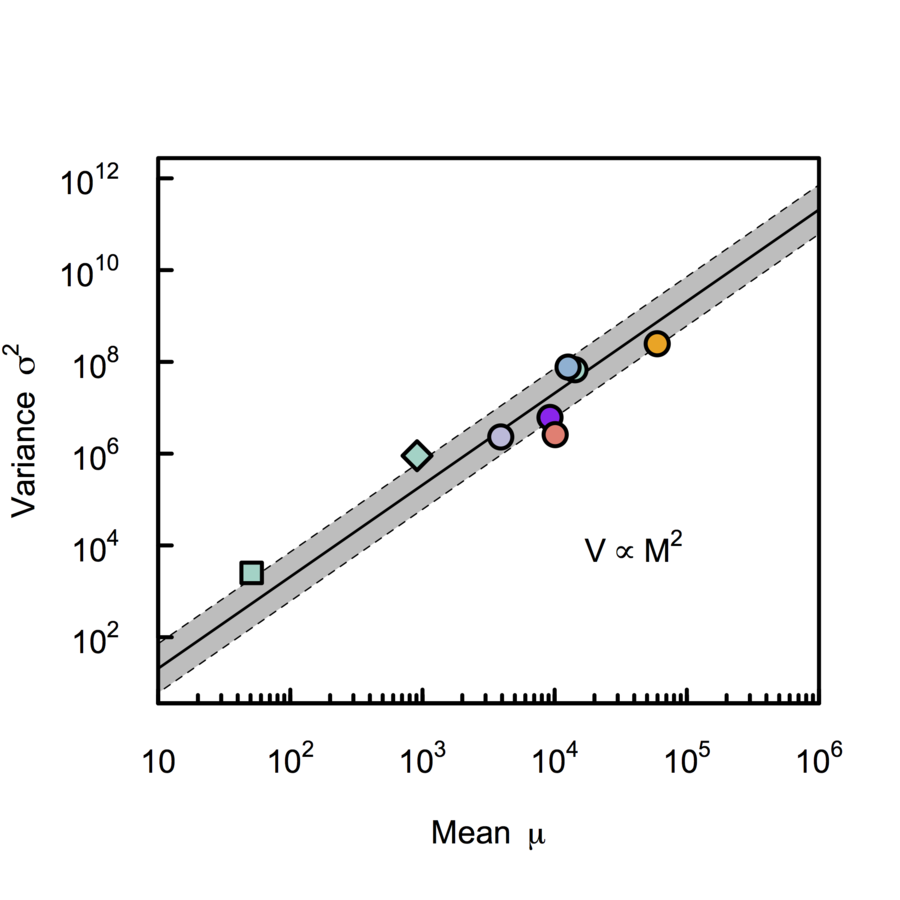
Cowan, (1984), Azevedo & Leroi (2000) PNAS




Data you shouldn't
go to war on
Data you do
go to war on
Thank you!
scRNA-seq
Gene specificity
about ~24'000 genes expressed in the brain.
Half-time2
By Daniel Fürth
Half-time2
- 504


