PyMKS Scaling
Daniel Wheeler, Berkay Yucel
PyMKS/Graspi Integration Meeting, 09/06/2021
Overview
- Investigate:
- PCA only speed up
- Full PCA pipeline speed up
- Full PCA pipeline memory usage
- Parameters:
- Samples
- Chunks
- Workers ???
- Outputs:
- Run time / speed up
- Memory Usage
- Accuracy (haven't instrumented yet)
Data
- Synthetic binary microstructures
- 8900 samples of 51x51x51
- Volume fraction from 25% to 75%
- 4 categories
Pipeline
def get_model():
return Pipeline([
('reshape', GenericTransformer(
lambda x: x.reshape(x.shape[0], 51, 51,51)
)),
('discritize', PrimitiveTransformer(n_state=2, min_=0.0, max_=1.0)),
('correlations', TwoPointCorrelation(periodic_boundary=True, correlations=[(0, 0)])),
('flatten', GenericTransformer(lambda x: x.reshape(x.shape[0], -1))),
('pca', PCA(n_components=3, svd_solver='randomized')),
('poly', PolynomialFeatures(degree=4)),
('regressor', LinearRegression(solver_kwargs={"normalize":False}))
])- All steps are Dask ML components
Preprocess Data
- Rechunk data in separate process
def prepare_data(n_sample, n_chunk):
x_data = da.from_zarr("../notebooks/x_data.zarr" , chunks=(100, -1))
x_data = x_data[:n_sample].rechunk((n_sample // n_chunk,) + x_data.shape[1:])
x_data.to_zarr('x_data.zarr', overwrite=True)Graphs and Chunks

Single Chunk
20 Chunks
Considerations
- Data is stored perfectly chunked for job using zarr (not always the case)
- Reality generally requires rechunking
- PCA is getting (n_sample, 123651) shaped arrays from 2 point stats (long, skinny). Current requirement for parallel PCA to be accurate and efficient
- Don't trust Dask linear regression currently, we have to look into that
- Only single node thus far (slurm cluster and laptop)
- Maxing out chunks to workers ratio may be bad for run times, only just realized this, needs to be investigated
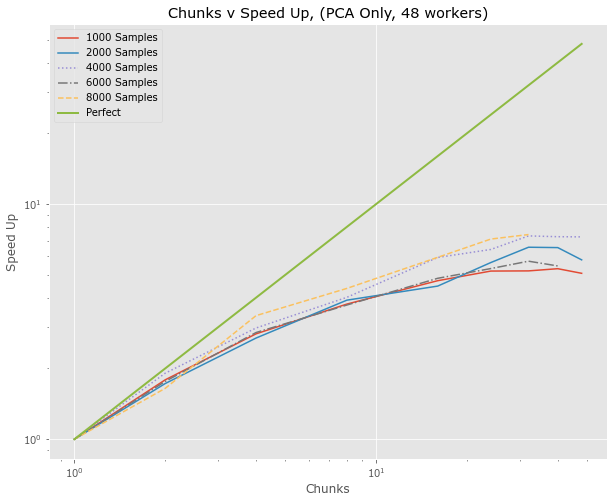
PCA Only
- Test the PCA Only as this is one of the main sources of communication
- 1000, 2000, 4000 on "rack3" node
- 6000, 8000 on "rack4" node (slower)
- Nodes have 128 total threads
- Would speed up be better or worse with shape of input (n_sample, n_feature)
- Maybe need more workers than 48
- Results are best of 5
PCA Only
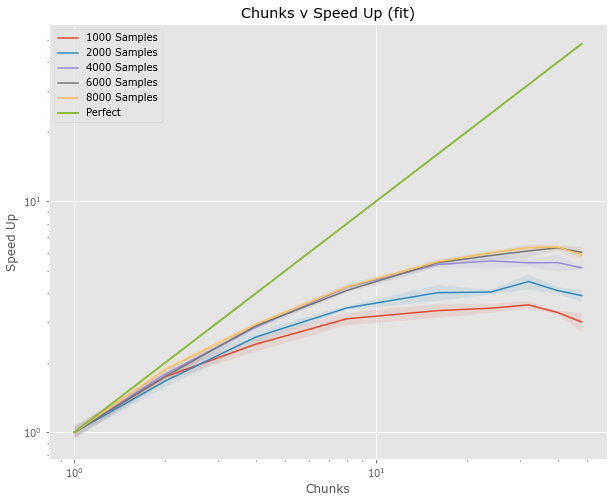
Full Pipeline (fit)
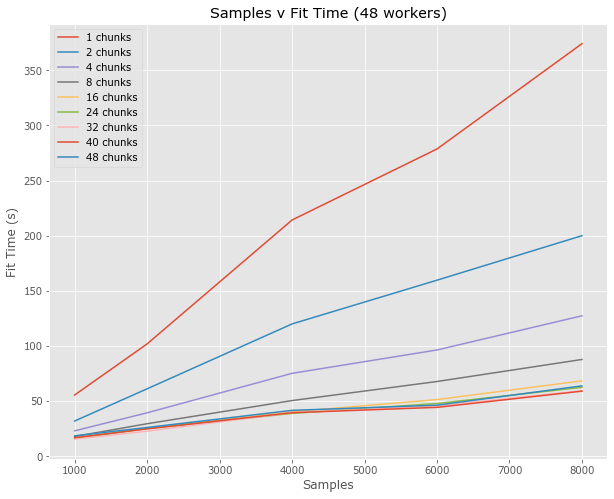
Full Pipeline (fit)
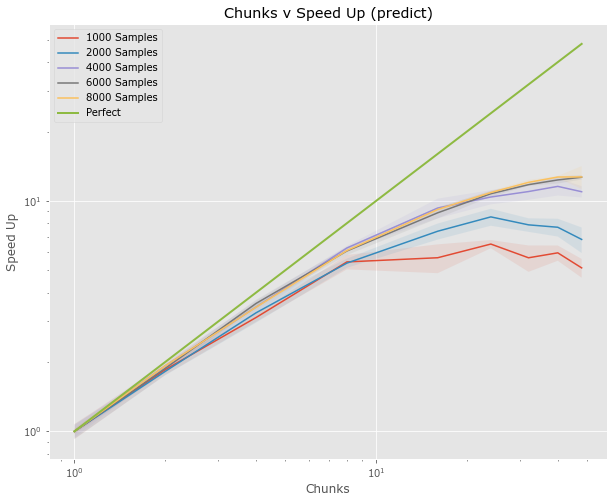
- Examine both fit and predict
- Results are median from 5
- Only 48 workers???
Full Pipeline (predict)
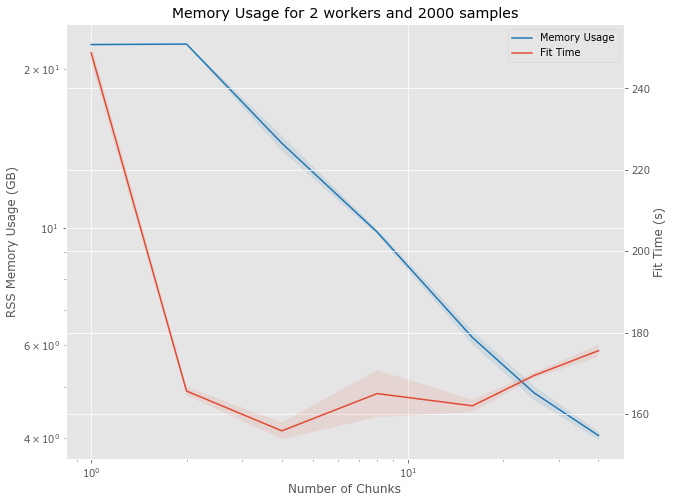
Memory Usage
- Following is on my laptop with 64 GB
- Many issues
- Collecting data on Slurm cluster
- Care must be taken with the final reduce operation in the MapReduce pipeline
- Only looking at 2000 samples of 51x51x51 data
- Uses 23GB with a single chunk
- Looking at 1 to 40 chunks
- Not considering accuracy and there will be an impact on accuracy.
- Median from 5 runs
- Pipeline includes reading data
Memory Usage
PyMKS Scaling
By Daniel Wheeler
PyMKS Scaling
- 624



